name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/transcriptomics" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/transcriptomics/tutorials/rna-seq-analysis-with-askomics-it/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" class="cover-logo"/> <br/> <br/> # Integrate and query local datasets and distant RDF data with AskOmics using Semantic Web technologies <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/xgaia/" class="contributor-badge contributor-xgaia"><img src="https://avatars.githubusercontent.com/xgaia?s=36" alt="Xavier Garnier avatar" width="36" class="avatar" /> Xavier Garnier</a> <li> <a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/abretaud?s=36" alt="Anthony Bretaudeau avatar" width="36" class="avatar" /> Anthony Bretaudeau</a> <li> <a href="/training-material/hall-of-fame/annesiegel/" class="contributor-badge contributor-annesiegel"><img src="https://avatars.githubusercontent.com/annesiegel?s=36" alt="Anne Siegel avatar" width="36" class="avatar" /> Anne Siegel</a> <li> <a href="/training-material/hall-of-fame/odameron/" class="contributor-badge contributor-odameron"><img src="https://avatars.githubusercontent.com/odameron?s=36" alt="Olivier Dameron avatar" width="36" class="avatar" /> Olivier Dameron</a> <li> <a href="/training-material/hall-of-fame/mboudet/" class="contributor-badge contributor-mboudet"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/mboudet?s=36" alt="Mateo Boudet avatar" width="36" class="avatar" /> Mateo Boudet</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00097">gxy.io/GTN:S00097</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Introduction to Galaxy Analyses](/training-material/topics/introduction) - [Sequence analysis](/training-material/topics/sequence-analysis) - Quality Control: [<i class="fab fa-slideshare" aria-hidden="true"></i><span class="visually-hidden">slides</span> slides](/training-material/topics/sequence-analysis/tutorials/quality-control/slides.html) - [<i class="fas fa-laptop" aria-hidden="true"></i><span class="visually-hidden">tutorial</span> hands-on](/training-material/topics/sequence-analysis/tutorials/quality-control/tutorial.html) - Mapping: [<i class="fab fa-slideshare" aria-hidden="true"></i><span class="visually-hidden">slides</span> slides](/training-material/topics/sequence-analysis/tutorials/mapping/slides.html) - [<i class="fas fa-laptop" aria-hidden="true"></i><span class="visually-hidden">tutorial</span> hands-on](/training-material/topics/sequence-analysis/tutorials/mapping/tutorial.html) --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - What is the Semantic Web and how can it help integrating and querying data? - How can AskOmics help benefiting from Semantic Web technologies without having to write RDF or SPARQL code? --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Understand the basics of RDF and SPARQL - Learn how input data must be structured to be integrated with AskOmics - Learn how to connect distant SPARQL endpoints to local data with AskOmics --- # How to explore data --- ## Requirements Study of biological mechanisms requires to: - **integrate** multiple data sources (differential expression results, genome annotation, remote protein database) - **query** them to answer a biological question (which genes are over-expressed in a condition, and what are the proteins coded by these genes) .image-00[ 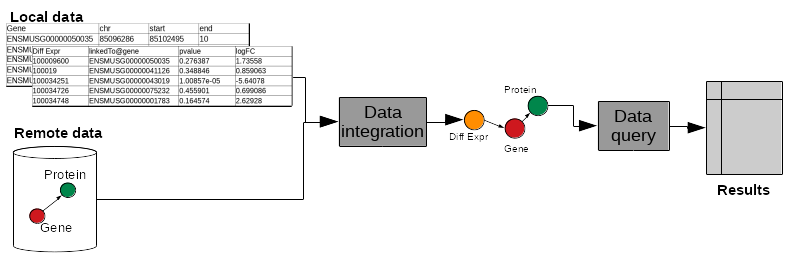] --- # What is the Semantic Web? --- ## Semantic Web Set of recommendations to **integrate data**, to **integrate domain knowledge** and to **perform query** and reasoning. - Resource Description Framework (RDF): annotate data - RDFS + OWL: represent knowledge (data description) - SPARQL Protocol and RDF Query Language (SPARQL): query data --- ## RDF - RDF is for describing **resources** (the R in RDF) - resources are identified by URIs (`nextprot:P01137`, `taxon:9606`) - **describing** (D in RDF) a resource is representing it explicitly - its attributes (`nextprot:P01137 :hasSequence "MPPSGLRLLL..."`) - its relations to other entities (`nextprot:P01137 :hasTaxon taxon:9606`) - its descriptions (aka classes) (`nextprot:P01137 :is nextprot:Protein`) --- ## RDF: Set of triples - RDF is represented by triples (subject, predicate, object) - **Subject**: the resource being described - **Predicate**: the relation (from subject to object) - **Object**: a value of the predicate for the subject .image-01[ ] ```turtle nextprot:P01137 :hasTaxon taxon:9606 . nextprot:P01137 :hasSequence "MPPSGLRLLL" . ``` --- ## RDF: triples form a labeled directed graph ```turtle # Description nextprot:P01137 rdf:type nextprot:Protein . taxon:9606 rdf:type nextprot:Organism . # Data nextprot:P01137 :hasTaxon taxon:9606 . nextprot:P01137 :hasSequence "MPPSGLRLLL" . ``` .image-02[ 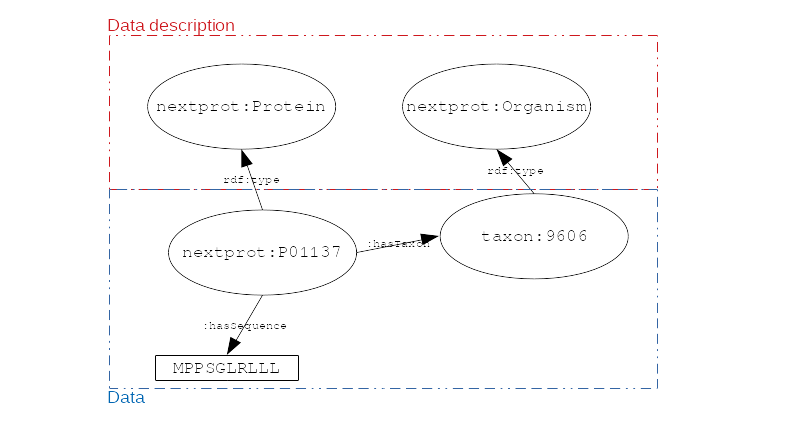] --- ## SPARQL - The SPARQL language is a set of triple patterns with variables (`?variable_name`) ``` SELECT ?gene WHERE { ?gene rdf:type :Gene . ?gene :hasTaxon taxon:9606 . } ``` - All `?gene` with `rdf:type` `:Gene` **and** with `:hasTaxon` `taxon:9606` - In other words, the query returns all the human genes --- ## SPARQL: entity matching allow federated queries .pull-left[ - Using the same identifier for the same entity (**entity matching**) across multiple datasets allows federated queries - Federated SPARQL queries provide unified querying capabilities over multiple datasets as if they were a single virtual graph ] .pull-right[ .image-03[ 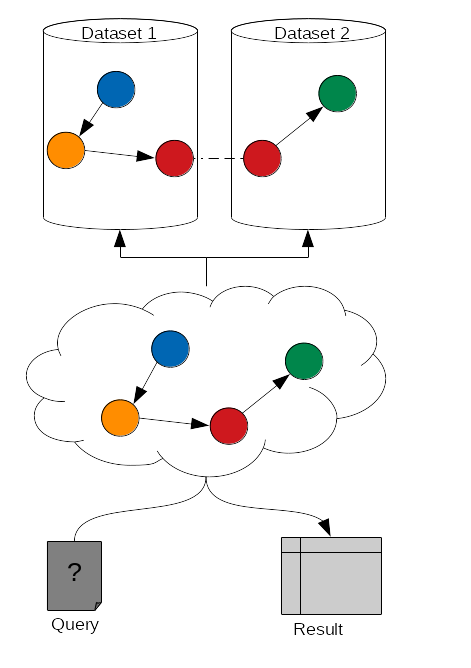] ] --- # What is AskOmics? --- ## AskOmics Web software for **data integration** and **query** using Semantic Web. The main functionalities are: - Convert of **multiple data formats** into RDF triples and store them in a local triplestore - Generate **complex SPARQL queries** using a user-friendly interface - Support external SPARQL endpoint to **cross-reference integrated data with remote data** AskOmics can be used as a standalone software, or **with Galaxy** --- # Data integration with AskOmics --- ## Local data integration - RDF and SPARQL are good infrastructures to describe and query biological datasets, but most of them are still stored on flat files like CSV/TSV and GFF. - AskOmics converts multiple structured data formats into RDF triples * CSV/TSV * GFF * BED --- ## AskOmics generates the graph of data and the abstraction AskOmics uses the file structure (*e.g.* header of TSV files) to generate the graph of data description: **the abstraction** .image-04[ 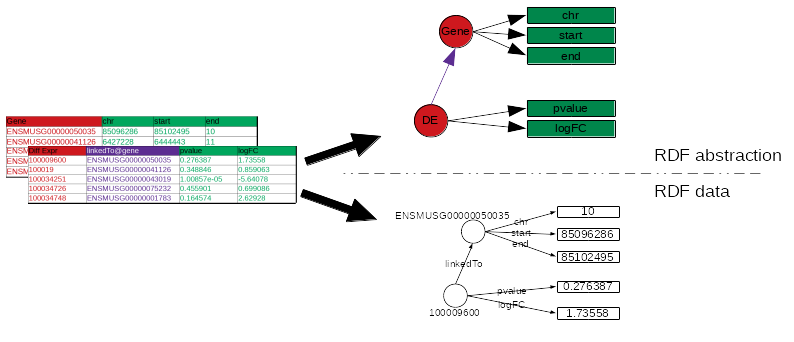] The rest of the files is converted to RDF triples that correspond to the data. --- ## Distant RDF data integration - Some public databases (*e.g.* [neXtProt](https://sparql.nextprot.org)) provide RDF data through a SPARQL endpoint (public access for RDF data) - To connect with a remote SPARQL endpoint, AskOmics needs its RDF abstraction - The abstraction can be generated with [abstractor](https://github.com/askomics/abstractor) ```bash pip3 install abstractor abstractor -s https://sparql.nextprot.org/sparql -o nextprot_abstraction.ttl -f turtle ``` - This abstraction can then be uploaded into AskOmics as a standard file --- # Query multiple data sources with AskOmics --- ## Query composition - Users navigate through the abstraction of local and remote data and create a path that represents a query .image-06[ ] - The query is converted to SPARQL code that is executed on the local and remote RDF data - Results are returned and can be downloaded --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - RDF and SPARQL are Semantic Web technologies that come useful for data integration and querying but the technical aspects may deter end-users - AskOmics is a web platform for data integration and query using Semantic Web in a user-friendly way --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/xgaia/" class="contributor-badge contributor-xgaia"><img src="https://avatars.githubusercontent.com/xgaia?s=36" alt="Xavier Garnier avatar" width="36" class="avatar" /> Xavier Garnier</a><a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-abretaud"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/abretaud?s=36" alt="Anthony Bretaudeau avatar" width="36" class="avatar" /> Anthony Bretaudeau</a><a href="/training-material/hall-of-fame/annesiegel/" class="contributor-badge contributor-annesiegel"><img src="https://avatars.githubusercontent.com/annesiegel?s=36" alt="Anne Siegel avatar" width="36" class="avatar" /> Anne Siegel</a><a href="/training-material/hall-of-fame/odameron/" class="contributor-badge contributor-odameron"><img src="https://avatars.githubusercontent.com/odameron?s=36" alt="Olivier Dameron avatar" width="36" class="avatar" /> Olivier Dameron</a><a href="/training-material/hall-of-fame/mboudet/" class="contributor-badge contributor-mboudet"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/mboudet?s=36" alt="Mateo Boudet avatar" width="36" class="avatar" /> Mateo Boudet</a> </td> </tr> <tr class="reviewers"> <td><abbr title="These people reviewed this material for accuracy and correctness">Reviewers</abbr></td> <td> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-badge-small contributor-hexylena"><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/xgaia/" class="contributor-badge contributor-badge-small contributor-xgaia"><img src="https://avatars.githubusercontent.com/xgaia?s=36" alt="Xavier Garnier avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-badge-small contributor-abretaud"><img src="https://avatars.githubusercontent.com/abretaud?s=36" alt="Anthony Bretaudeau avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-badge-small contributor-gallardoalba"><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-badge-small contributor-shiltemann"><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /></a></td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>