name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/visualisation" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/visualisation/tutorials/jbrowse/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" class="cover-logo"/> <br/> <br/> # JBrowse <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /> Helena Rasche</a> <li> <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /> Saskia Hiltemann</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00102">gxy.io/GTN:S00102</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- # JBrowse for Genomics Visualisation --- ### What is JBrowse .pull-left[ - Interactive genome browser - HTML5 / Javascript - Process data + copy directory to a webserver - Can be deployed to apache/nginx, S3 buckets, etc. - Many [plugins](https://gmod.github.io/jbrowse-registry/) available for - Themes - Datatype support (mafviewer, agp_parser) - Third party data integration (GA4GH, MyVariantViewer) - Visualisations (GC Content track, Sashimi for RNAseq) - More ] .pull-right[ 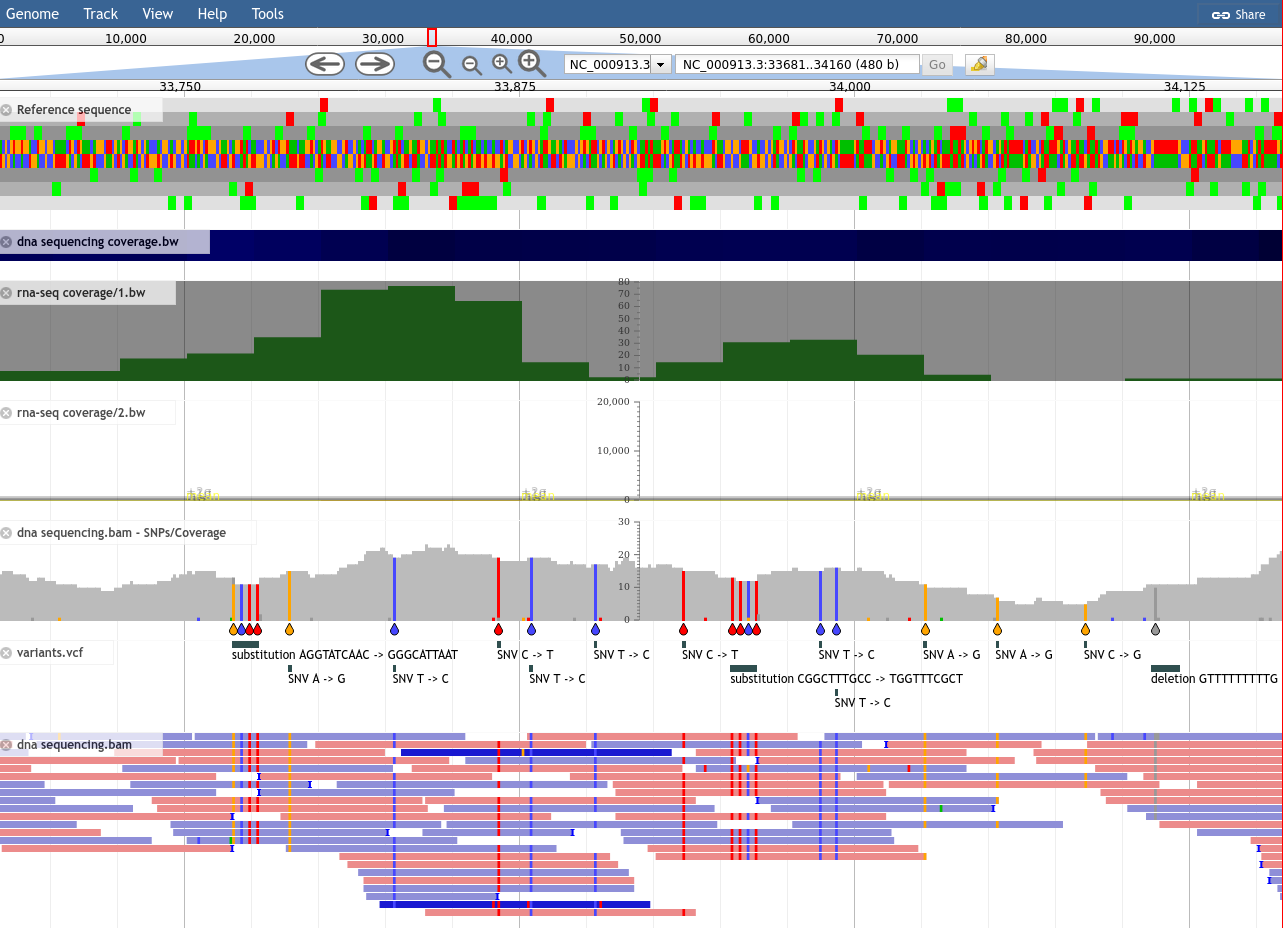 ] --- ### JBrowse in Galaxy - Tool that produces a JBrowse instance (client side code + processed data) - Workflow compatible - Covers *most* of the possibilities of JBrowse - Supports many Galaxy datatypes: - GFF3 / GBK /BED - BAM (SAM) - (big)Wig - Blast XML - VCF - REST/SPARQL endpoints - Complex JBrowse made easy (Thanks Galaxy!) --- ### Built in Provenance Tracking - "ComboTrackSelector" supports attaching metadata to tracks + filtering - We attached Galaxy metadata - Dataset Creator - Dataset Job Parameters - Tool + Version - Date - Can "build on top of" existing JBrowses, adding more data. Complete history tracked. 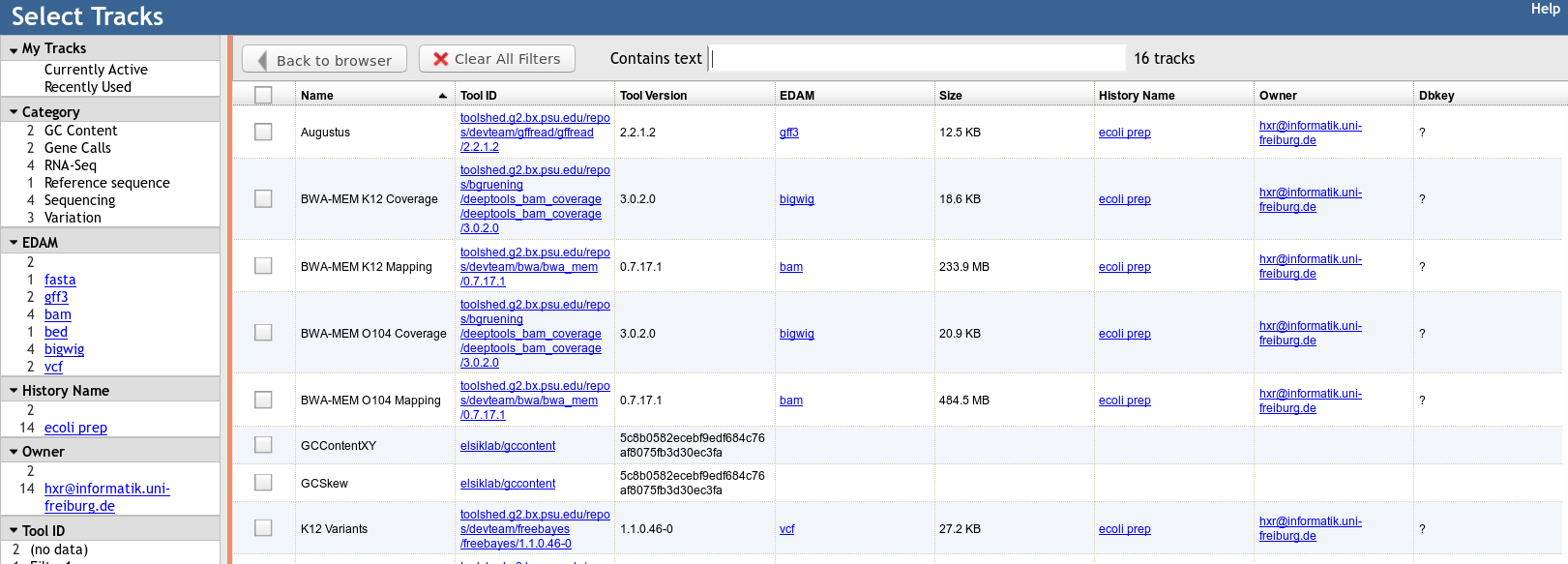 --- ### Why is this Necessary? - JBrowse allows you to present Galaxy analysis workflows in a single visualisation - Researchers generally understand genome browsers - When annotation, not just visualisation, is needed, the JBrowse instances can be sent to Apollo - Apollo allows for collaborative genome annotation and curation - Check out the Apollo tutorials for more details: [prokaryotes](/training-material/topics/genome-annotation/tutorials/apollo/tutorial.html) - [eukaryotes](/training-material/topics/genome-annotation/tutorials/apollo-euk/tutorial.html) --- ### Why do you need this? - You are running genome annotation or genomic data analysis workflows - Researchers need more visual outputs than tables of data - Datasets can be more valuable visualised together, rather than as separate lines in a report --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-hexylena"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /> Helena Rasche</a><a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /> Saskia Hiltemann</a> </td> </tr> <tr class="reviewers"> <td><abbr title="These people reviewed this material for accuracy and correctness">Reviewers</abbr></td> <td> <a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-badge-small contributor-bgruening"><img src="https://avatars.githubusercontent.com/bgruening?s=36" alt="Björn Grüning avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-badge-small contributor-abretaud"><img src="https://avatars.githubusercontent.com/abretaud?s=36" alt="Anthony Bretaudeau avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-badge-small contributor-hexylena"><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-badge-small contributor-shiltemann"><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /></a></td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" style="height: 100px;"/> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>