Intro to DataPLANT ARCs
Contributors
last_modification Published: Oct 24, 2024
last_modification Last Updated: Oct 28, 2024
About DataPLANT
Towards democratization of plant research.
Speaker Notes
- DataPLANT is a consortium from the heart of the German plant research community.
- It aims to establish sustainable Research Data management, RDM, by providing both digital assistance, such as software or teaching material, as well as and personal assistance, for example via on-site consultation or workshops.
- DataPLANT is committed to developing an RDM system that meets community requirements and facilitates the processing and contextualization of research datasets in accordance with the FAIR principles (Findable, Accessible, Interoperable, Reusable).
About DataPLANT
.pull-left[
- DataPLANT’s mission is to lead the digital transformation in plant science by advancing from traditional publications to innovative data-driven formats like Annotated Research Contexts (ARC).
- DataPLANT builds user-friendly services that simplify data annotation and metadata management for plant scientists. By leveraging existing IT infrastructure, it aims to make the process as seamless and efficient as possible.
]
.pull-right[
 ]
]
.footnote[ nfdi4plants.org] Speaker Notes
- DataPLANT’s mission is to lead the digital transformation in plant science by advancing from traditional publications to innovative data-driven formats like Annotated Research Contexts (ARC).
- DataPLANT builds user-friendly services that simplify data annotation and metadata management for plant scientists. By leveraging existing IT infrastructure, it aims to make the process as seamless and efficient as possible.
- You can read more about dataplant at nfdi4plants.org
Data Stewardship between DataPLANT and communities
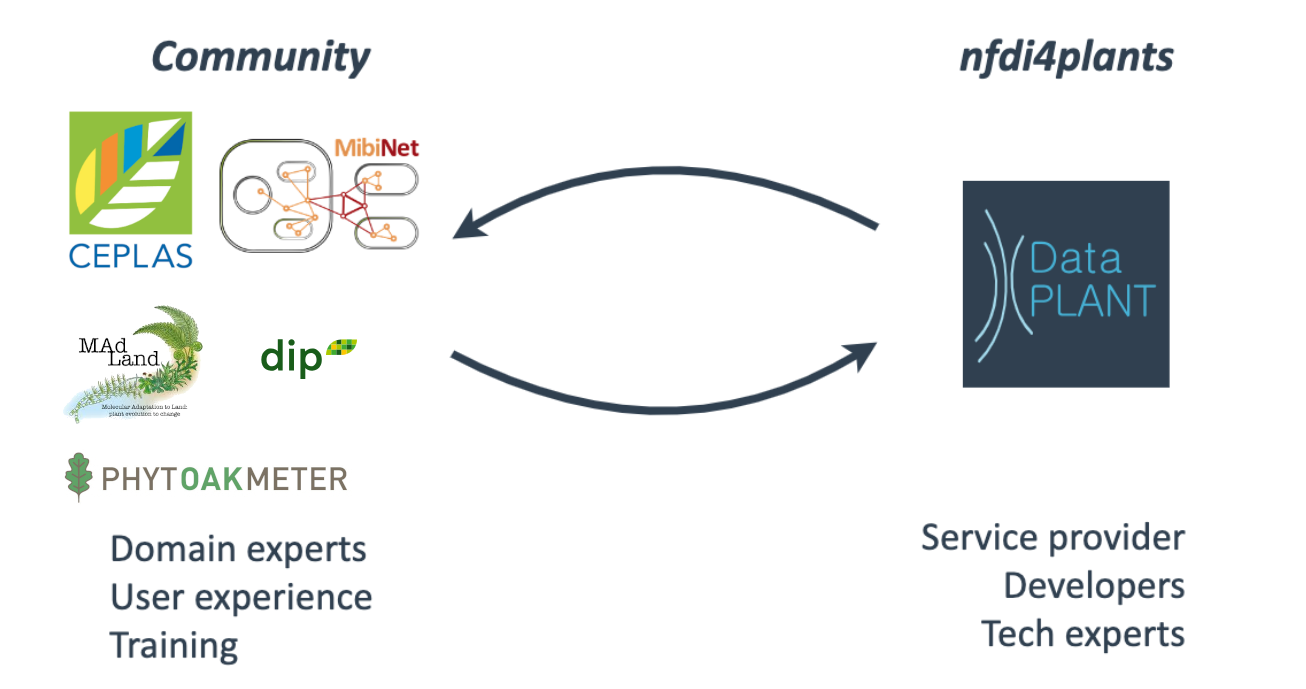
Speaker Notes
- DataPLANT works closely together with various plant consortia and projects.
- DataPLANT acts as the service provider, and has a team of technology experts and semantic specialists.
- DataPLANT supports communities through their tools, services and consultation.
- And in turn, the communities provide feedback and contributions to DataPLANT.
Annotated Research Context (ARC)
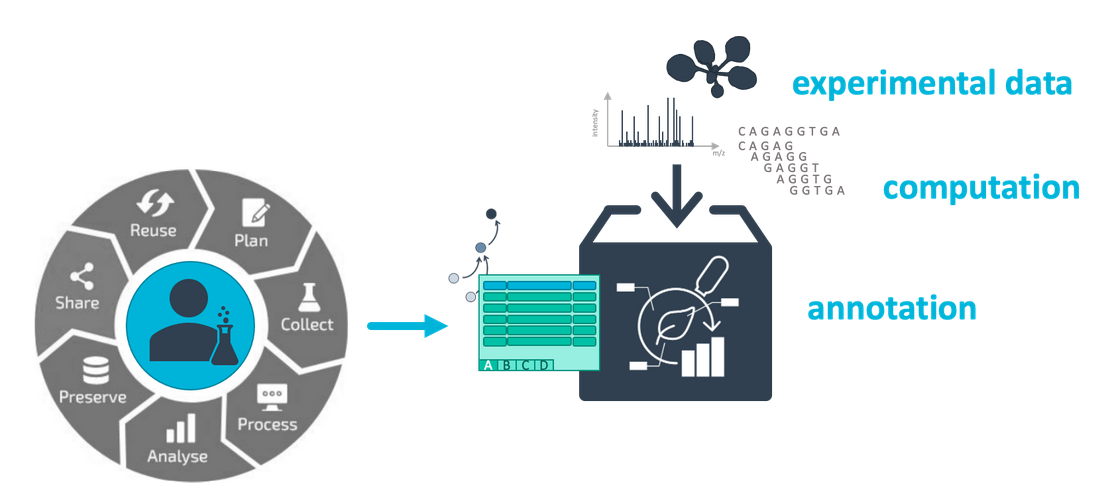
Your entire investigation in a single unified bag
Speaker Notes
- Annotated research contexts, or ARCs for short, provide a way to bundle your entire investigation in one unified place.
- ARCs can contain your experimental data and annotation, as well as your computational results and workflows.
- ARCs allow you to share your research in a FAIR and open way.
What does an ARC look like?
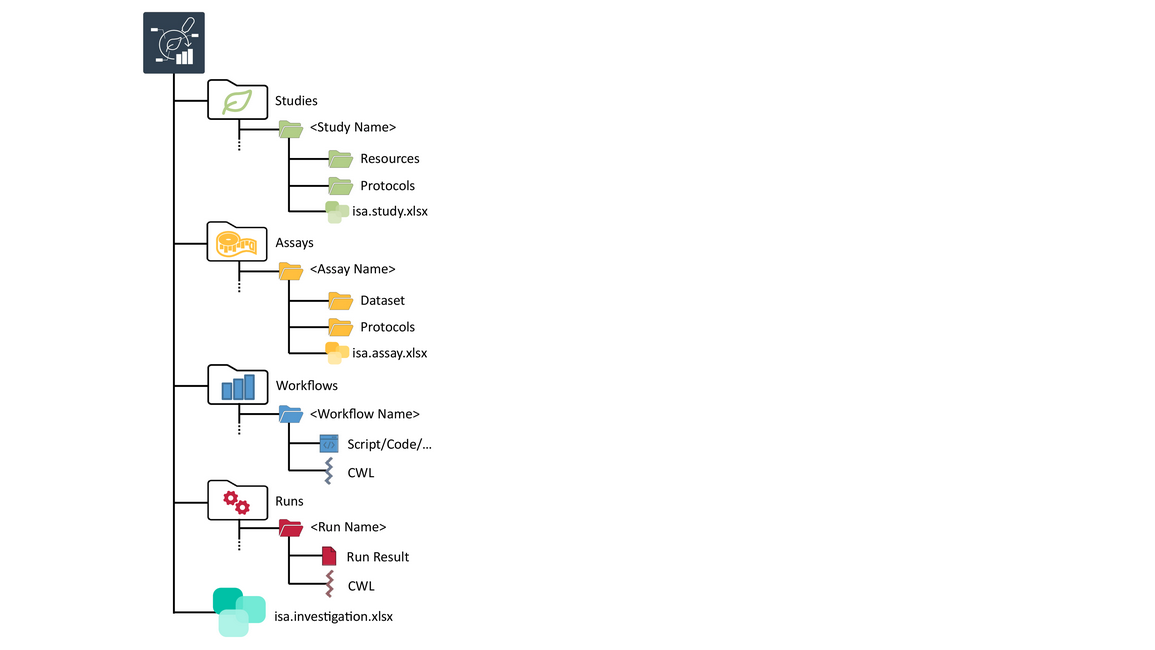
Speaker Notes
- An ARC, at its core, is a structured folder of data.
- This structure is based on the ISA data model.
- ISA stands for investigation, study, assay.
- Every ARC represents an investigation, and contains one or more studies, assays, workflows and runs at its root.
- We will focus mostly on studies and assays in this tutorial.
- This is where you put your experimental data, and where you usually start when creating your ARC.
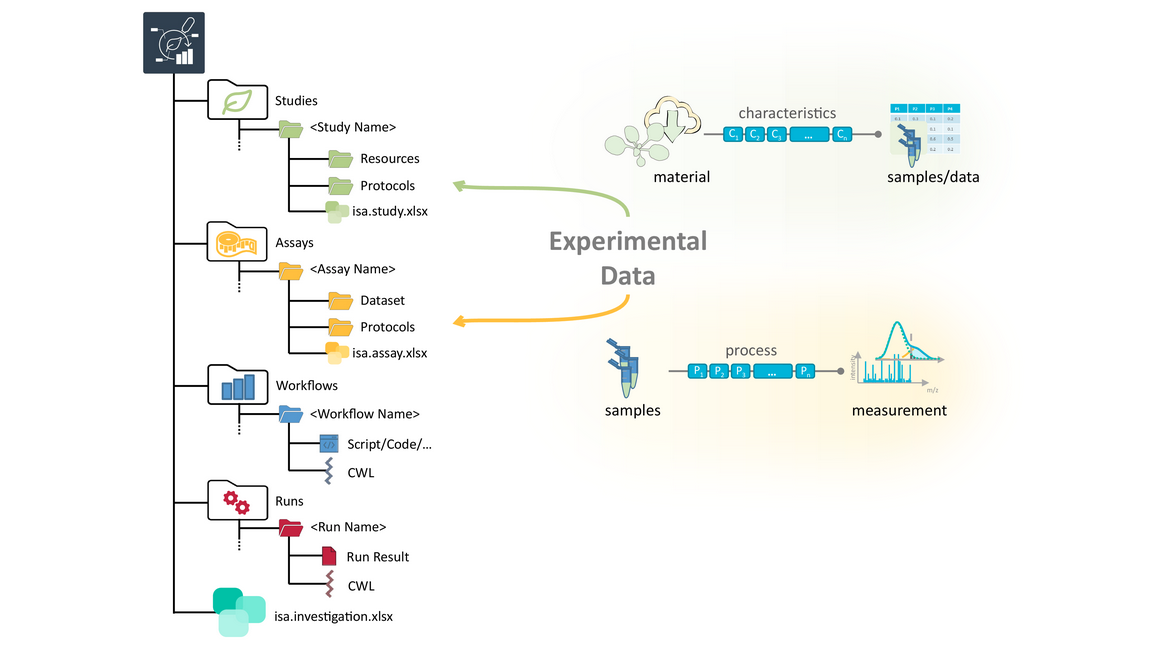
ARCs store experimental data
Speaker Notes
- Studies contain information about the biological materials you used in your research. The plants you grew, but also lab protocols chemicals you used.
- Assays contain results and metadata about any measurements you performed.
- At the end of a measurement you either have another sample, for example in the case of an extraction, or you have data, for example for a sequencing assay.
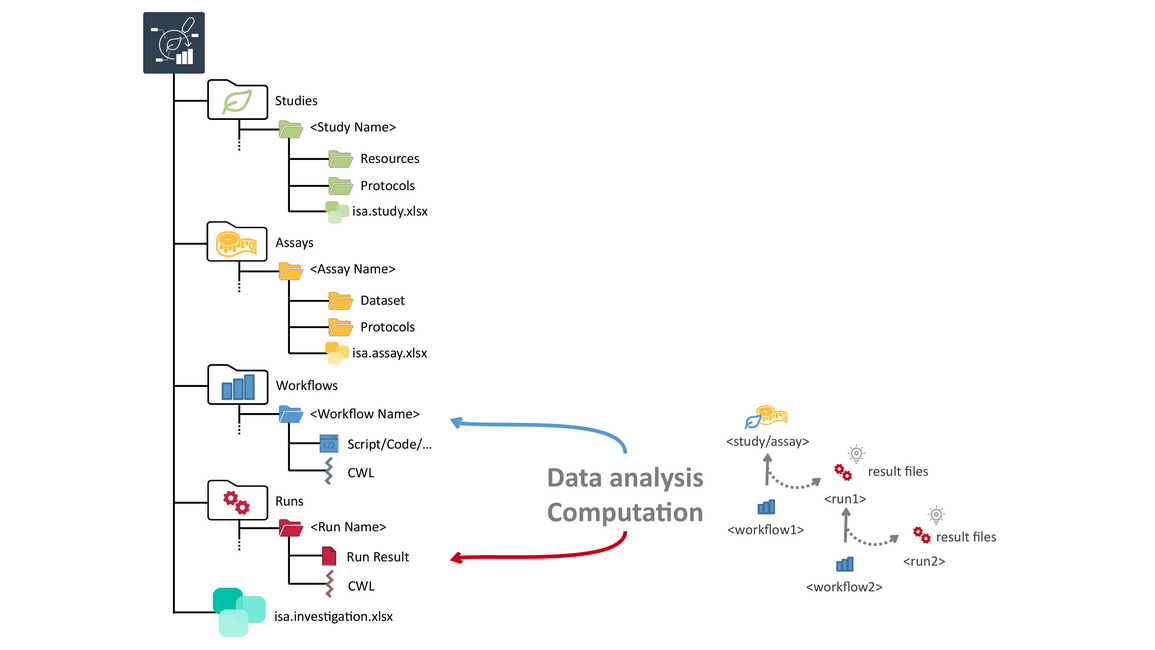
Computations can be run inside ARCs
Speaker Notes
- In the workflows folder you would store any scripts or workflows used to analyze the data coming from your assays.
- By specifying CWL workflows, your bioinformatics analysis can be reproduced, right inside the ARC.
- Any results from these analysis workflows are stored in the runs folder.
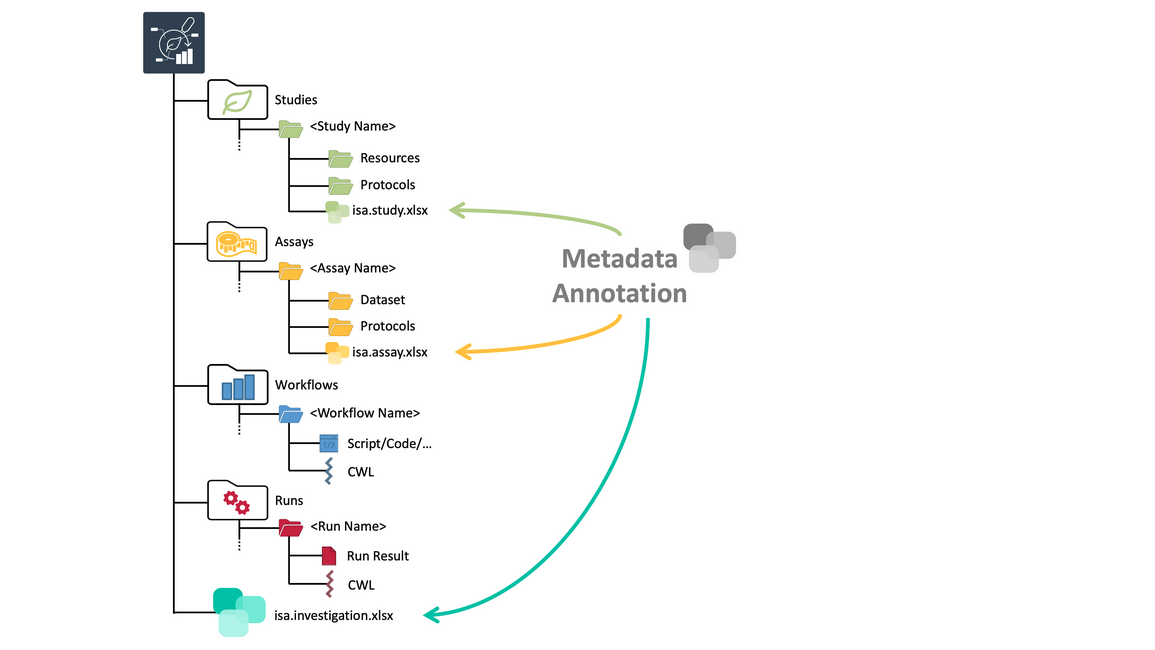
ARCs come with comprehensive metadata
Speaker Notes
- In addition to raw data, ARCs also contain structured metadata.
- This metadata uses ontologies to describe your research.
- Metadata annotations are stored in so-called ISA files. These are stored as excel workbooks in the ARC.
- There is investigation level metadata in the ISA investigation file.
- And similarly we have study-level and assay-level metadata files.
- For example, on the investigation level this is high-level information about your research, who you are, what your biological questions are, what the experimental design was, any related publications, etcetera.
- On the study level, you would describe things like your plant samples, how they were grown, harvested and cultured.
- On the assay level, you would describe information about the measurement.
- For example for a sequencing assay you would describe the RNA or DNA extraction, the library preparation and the instruments used. Essentially the aim is to capture the entire path of your samples in the lab.
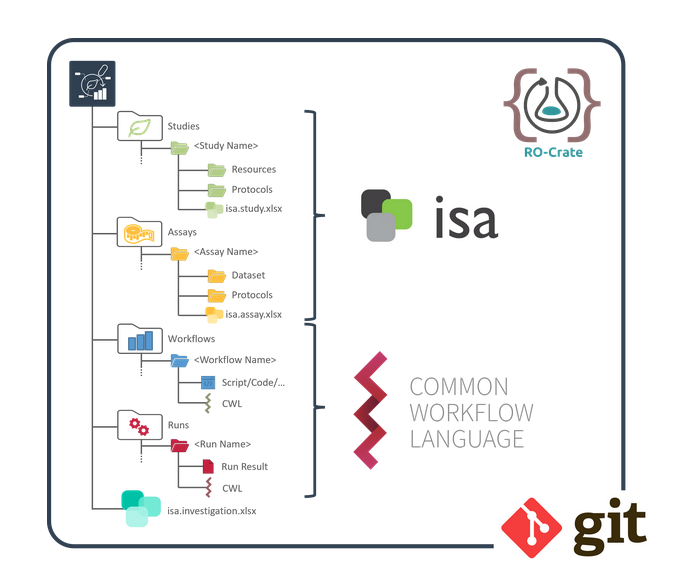
ARC builds on standards
.pull-left[
 ]
]
.pull-right[
</br>
ARC incorporate established standards
- RO-Crate: standardized exchange
- ISA: structured, machine-readable metadata
- CWL: reproducible, re-usable data analysis
- Git: version control
- Ontologies: standardized metadata ]
Speaker Notes
- All of this builds on existing standards.
- ARCs are an RO-crates implementation.
- They use the ISA data model.
- CWL is used to describe data analysis.
- Git is used for version control.
- Ontologies are leveraged to standardize metadata.
You can store ARCs in the DataHUB

Speaker Notes
- Typically you start creating your ARC on your computer.
- But you can store ARCs online in the DataHUB, and thereby also creating a backup of your research.
- You can make changes to your ARC locally on your computer, push it to DataHUB, and from there sync it again, maybe to a different computer.
ARCs are versioned
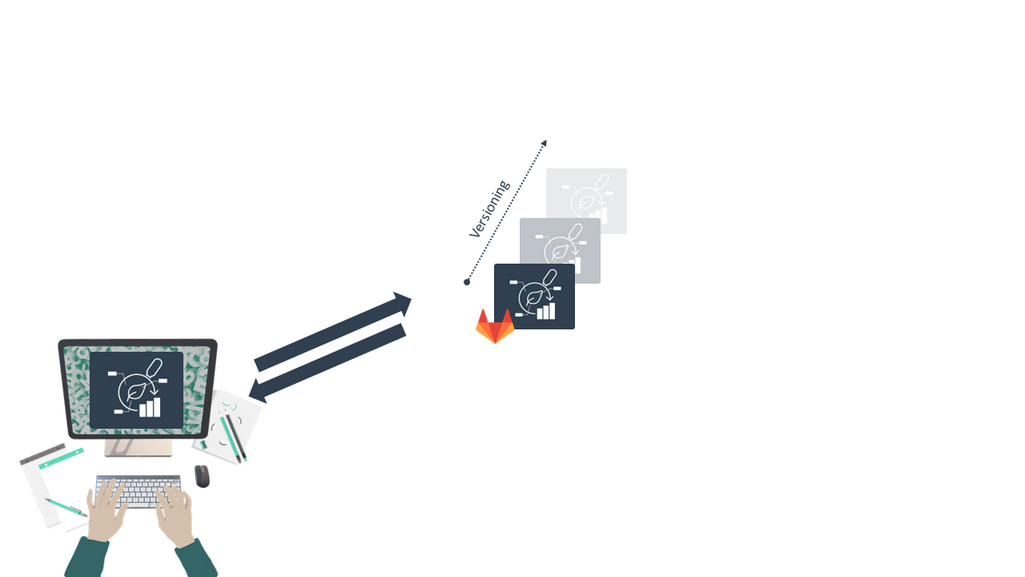
Speaker Notes
- DataHUB also provides version control for your ARC.
- This means you have a detailed log of how your ARC changed over time, and you can always go back to a previous version if needed.
You can invite collaborators
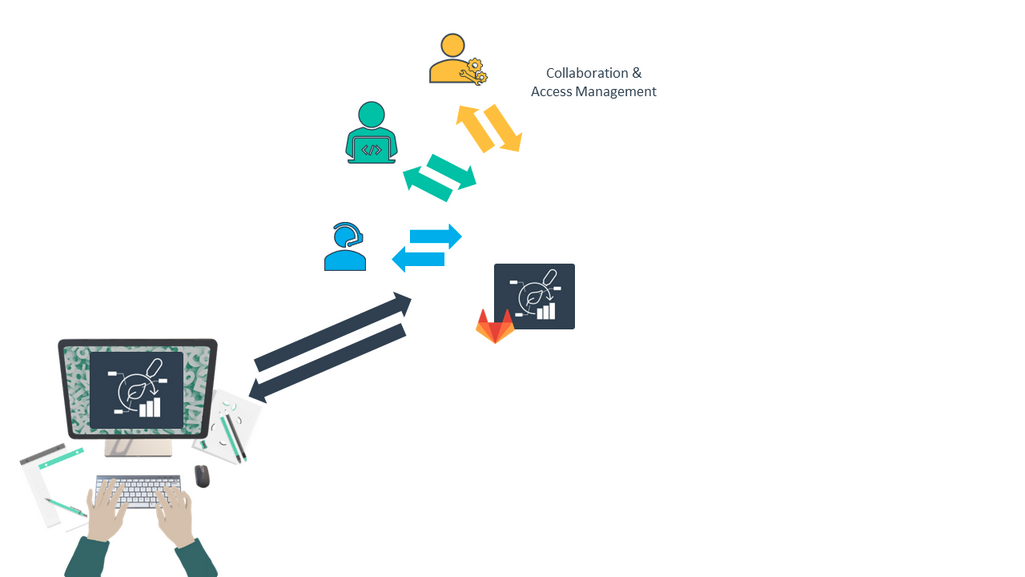
Speaker Notes
- By default your ARC is private to you on DataHUB.
- But you can invite other people to collaborate on your ARC, by giving them access to your ARC.
- This can be other people from your lab, or people from other institutes.
Collaborate and Contribute
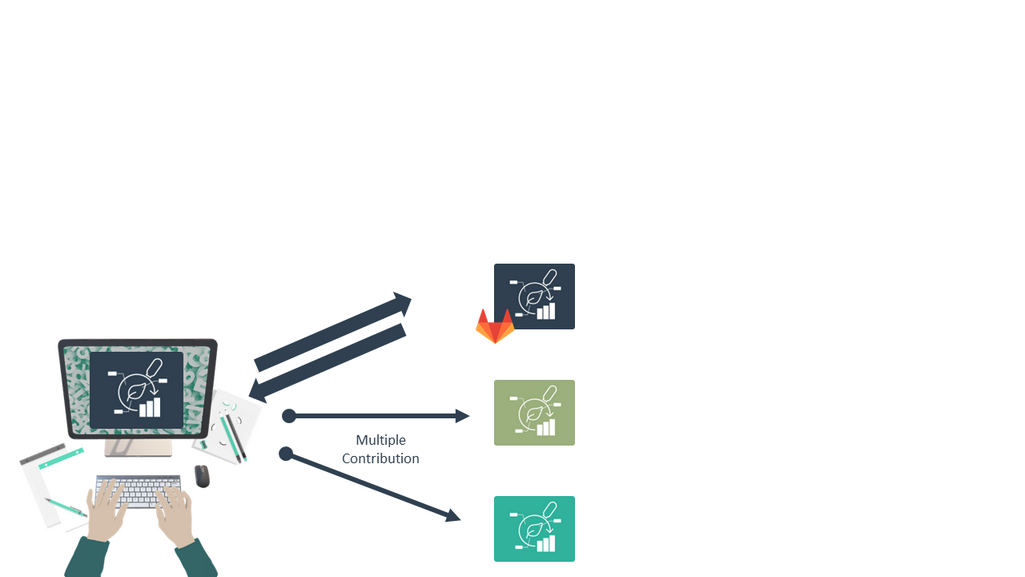
Speaker Notes
- You can contribute to multiple ARCs, multiple research projects.
- For example, if others invited you to collaborate, you can contribute to their ARC.
- Or if you have multiple research projects of your own, you can have multiple ARCs on DataHUB.
Reuse data in ARCs
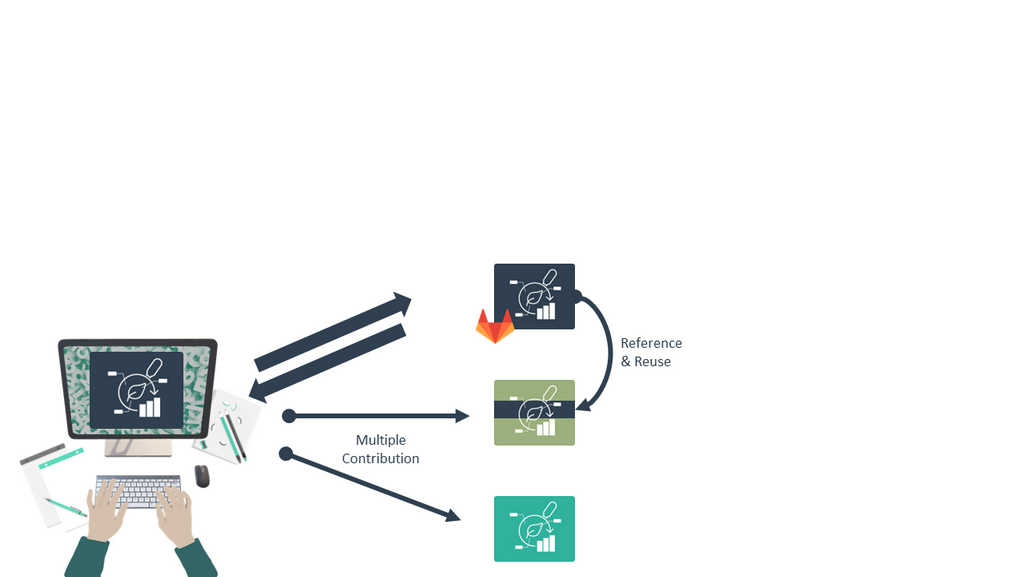
Speaker Notes
- You can also reuse parts of other ARCs, so you don’t always have to recreate things like scripts, protocols, assays, and other shared research components.
Publish your ARC
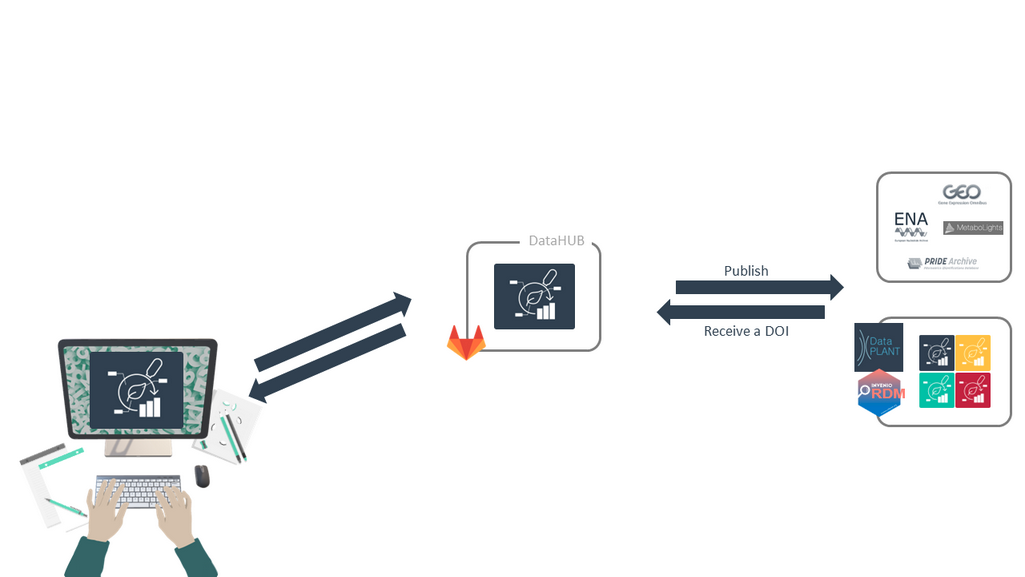
Speaker Notes
- Once your ARC is complete, and you are ready to release your work, you can publish your ARC.
- You will receive a DOI, a digital object identifier, for your ARC.
- DataPLANT is also creating converters for popular data repositories such as ENA, GEO, and NCBI.
- For example, if the editor of your journal requires you to deposit your data in one of these repositories, you can easily extract the data and information from your ARC in the appropriate format for these repositories.
Publish your ARC, get a DOI
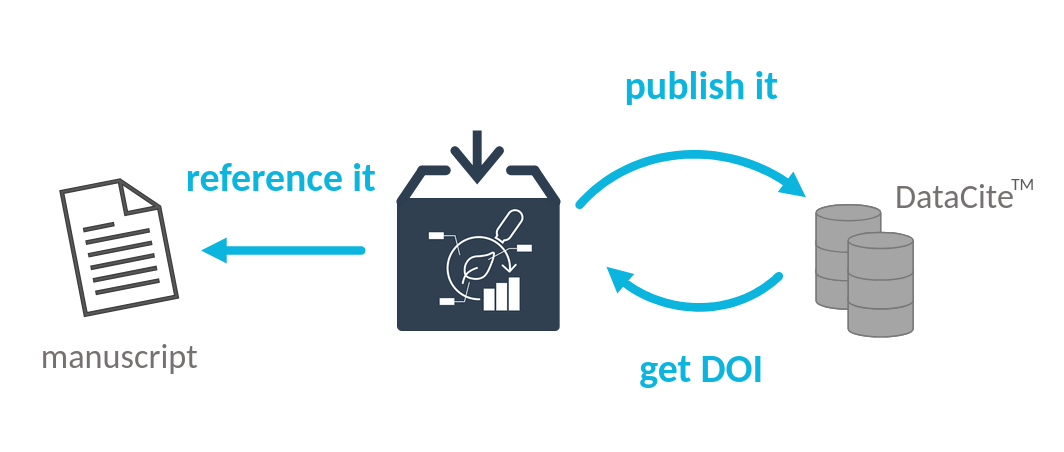
Speaker Notes
- The DOI you receive for your ARC can be referenced in your journal article, anabling readers to reuse your data and workflows.
- If you make changes to your ARC, you can publish a new version, and receive a new DOI, while your original DOI will always point to the original version of your ARC.
Moving from paper to data publications
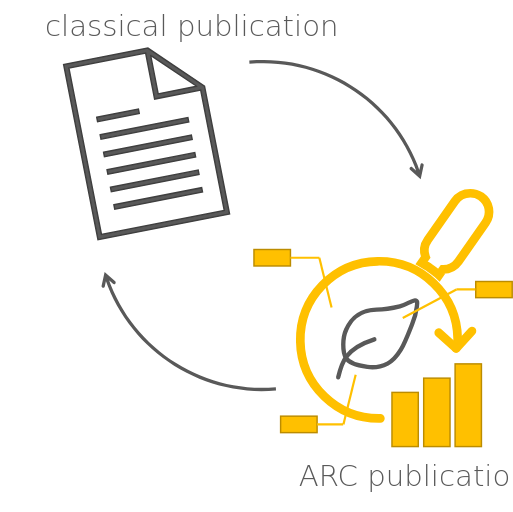
Speaker Notes
- This approach allows us to move from classical publications to a more data-centric publication model.
ARC ecosystem
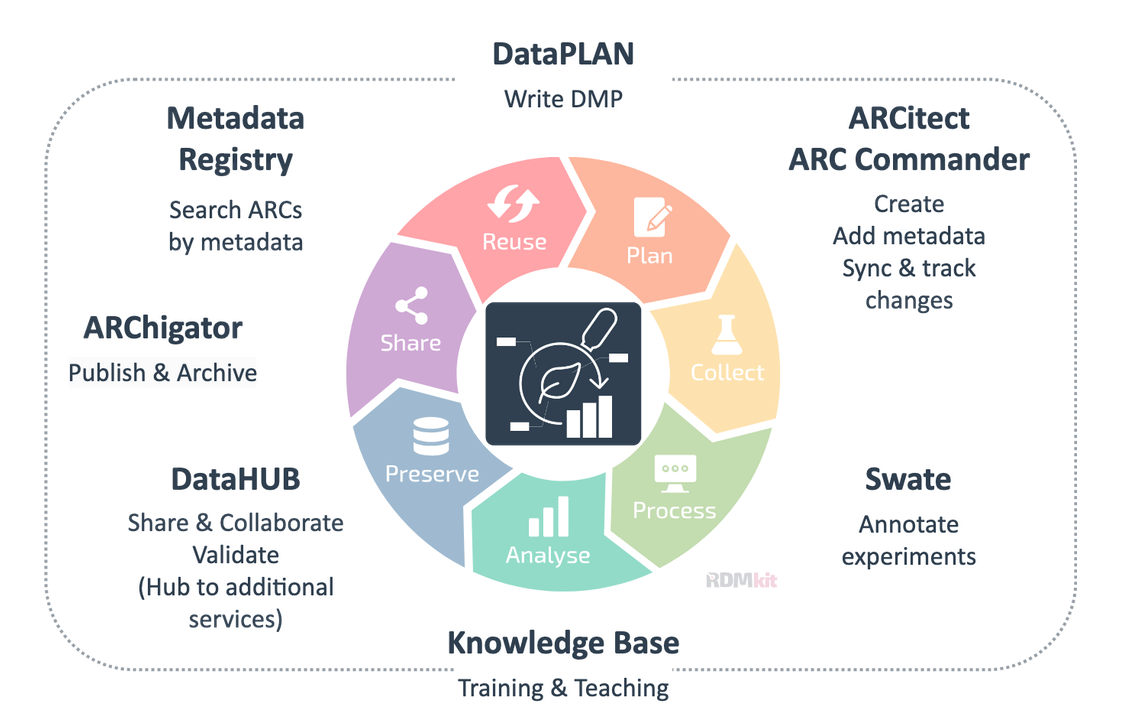
Speaker Notes
- DataPLANT offers an entire ecosystem of tools and services around this concept, in all phases of the research data management cycle.
- From writing your data management plan, to storing and describing your research data, sharing and collaborating, and finally publishing your research and making it findable and accessible to scientists worldwide.
Thank you!
This material is the result of a collaborative work. Thanks to the Galaxy Training Network and all the contributors! Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.
Tutorial Content is licensed under
Creative Commons Attribution 4.0 International License.