name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/fair" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/fair/tutorials/earth_system_rocrate/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" class="cover-logo"/> <br/> <br/> # An earth-system RO-Crate <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/Marie59/" class="contributor-badge contributor-Marie59"><img src="https://avatars.githubusercontent.com/Marie59?s=36" alt="Marie Josse avatar" width="36" class="avatar" /> Marie Josse</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00127">gxy.io/GTN:S00127</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="far fa-play-circle" aria-hidden="true"></i><span class="visually-hidden">video-slides</span> <a href="/training-material/videos/watch.html?v=/fair/tutorials/earth_system_rocrate/slides">Video slides</a> | <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - How to generate a RO-Crate - How to follow a Marine Omics study - How to read a RO-Crate ? --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Understand RO-Crate in an earth-system context --- # Workflow context The workflow used as an example here goes from : - First, produce a protein FASTA file from a nucleotide FASTA file using (<span class="tool" data-tool="tool_id=toolshed.g2.bx.psu.edu/repos/iuc/prodigal/prodigal/2.6.3+galaxy0" title="Prodigal tool" aria-role="button"><i class="fas fa-wrench" aria-hidden="true"></i> <strong>Prodigal</strong> (<i class="fas fa-cubes" aria-hidden="true"></i> Galaxy version 2.6.3+galaxy0)</span>) (a tool that predicts protein-coding genes from DNA sequences). - Then, use (<span class="tool" data-tool="toolshed.g2.bx.psu.edu/repos/bgruening/interproscan/interproscan/5.59-91.0+galaxy3" title="InterProscan tool" aria-role="button"><i class="fas fa-wrench" aria-hidden="true"></i> <strong>InterProscan</strong> (<i class="fas fa-cubes" aria-hidden="true"></i> Galaxy version 5.59-91.0+galaxy3)</span>) to create a tabular. Interproscan is a batch tool to query the InterPro database. It helps identify and predict the functions of proteins by comparing them to known databases. - Finally, discover (<span class="tool" data-tool="toolshed.g2.bx.psu.edu/repos/ecology/sanntis_marine/sanntis_marine/0.9.3.5+galaxy1" title="SanntiS tool" aria-role="button"><i class="fas fa-wrench" aria-hidden="true"></i> <strong>SanntiS</strong> (<i class="fas fa-cubes" aria-hidden="true"></i> Galaxy version 0.9.3.5+galaxy1)</span>) both to build genbank and especially to detect and annotate biosynthetic gene clusters (BGCs). --- ## The workflow itself <iframe title="Galaxy Workflow Embed" style="width: 100%; height: 700px; border: none;" src="https://earth-system.usegalaxy.eu/published/workflow?id=b9c938d1af08124b&embed=true&buttons=true&about=false&heading=false&minimap=true&zoom_controls=true&initialX=-20&initialY=-20&zoom=1"></iframe> --- ## Run the workflow Import and run the [Biosynthetic Gene Clusters workflow](https://earth-system.usegalaxy.eu/u/marie.josse/w/marine-omics-identifying-biosynthetic-gene-clusters) To know more on how to do that follow the [tutorial](/training-material/topics/ecology/tutorials/marine_omics_bgc/tutorial.html) --- ## Generate the RO-Crate Once you run the workflow you can generate the RO-Crate. To do so follow the [tutorial](/training-material/topics/fair/tutorials/ro-crate-in-galaxy/tutorial.html) --- ### From workflow to RO-Crate 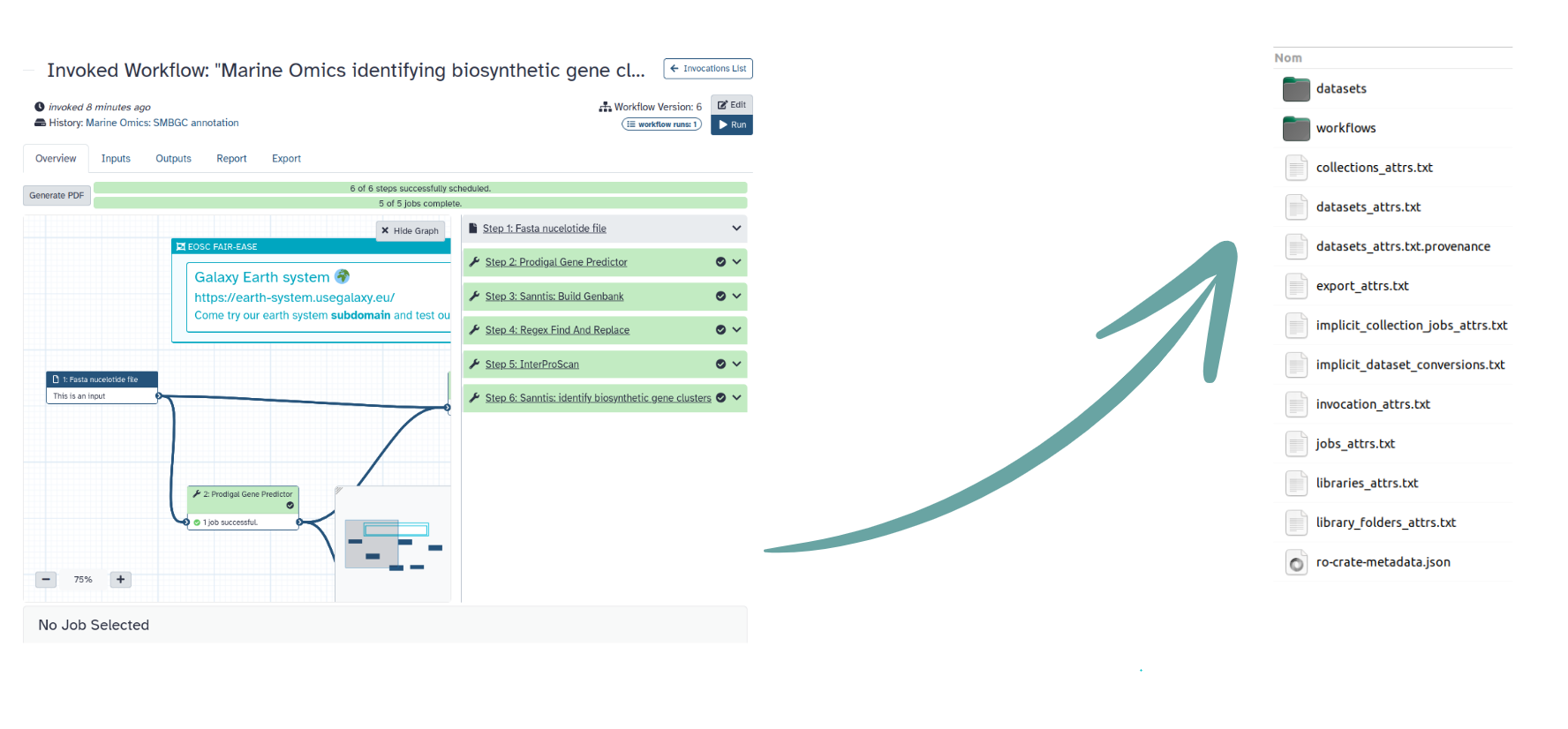 --- ### The RO-Crate folder and architecture 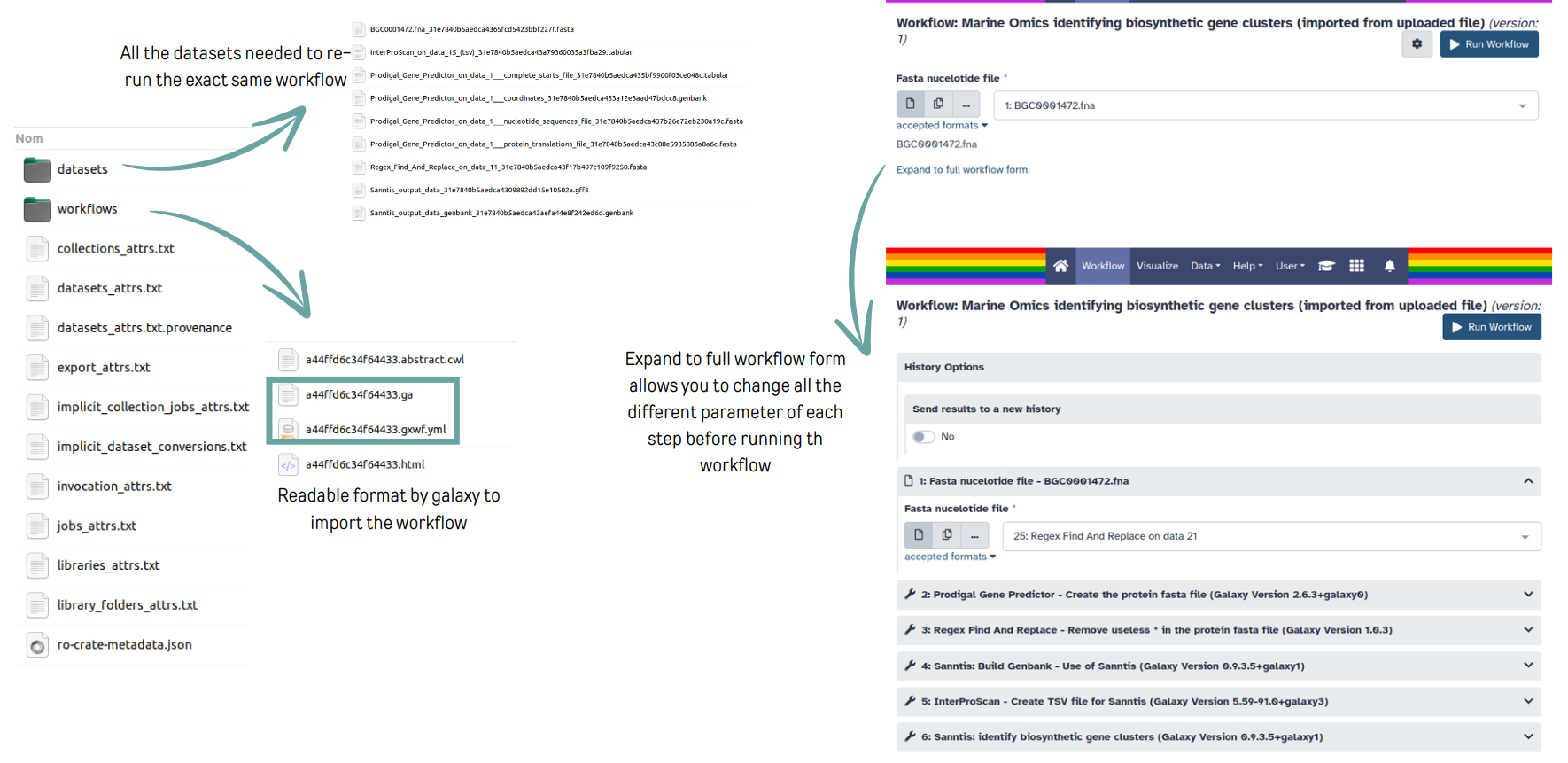 --- ### Let's dive into the ro-crate-metadata.json 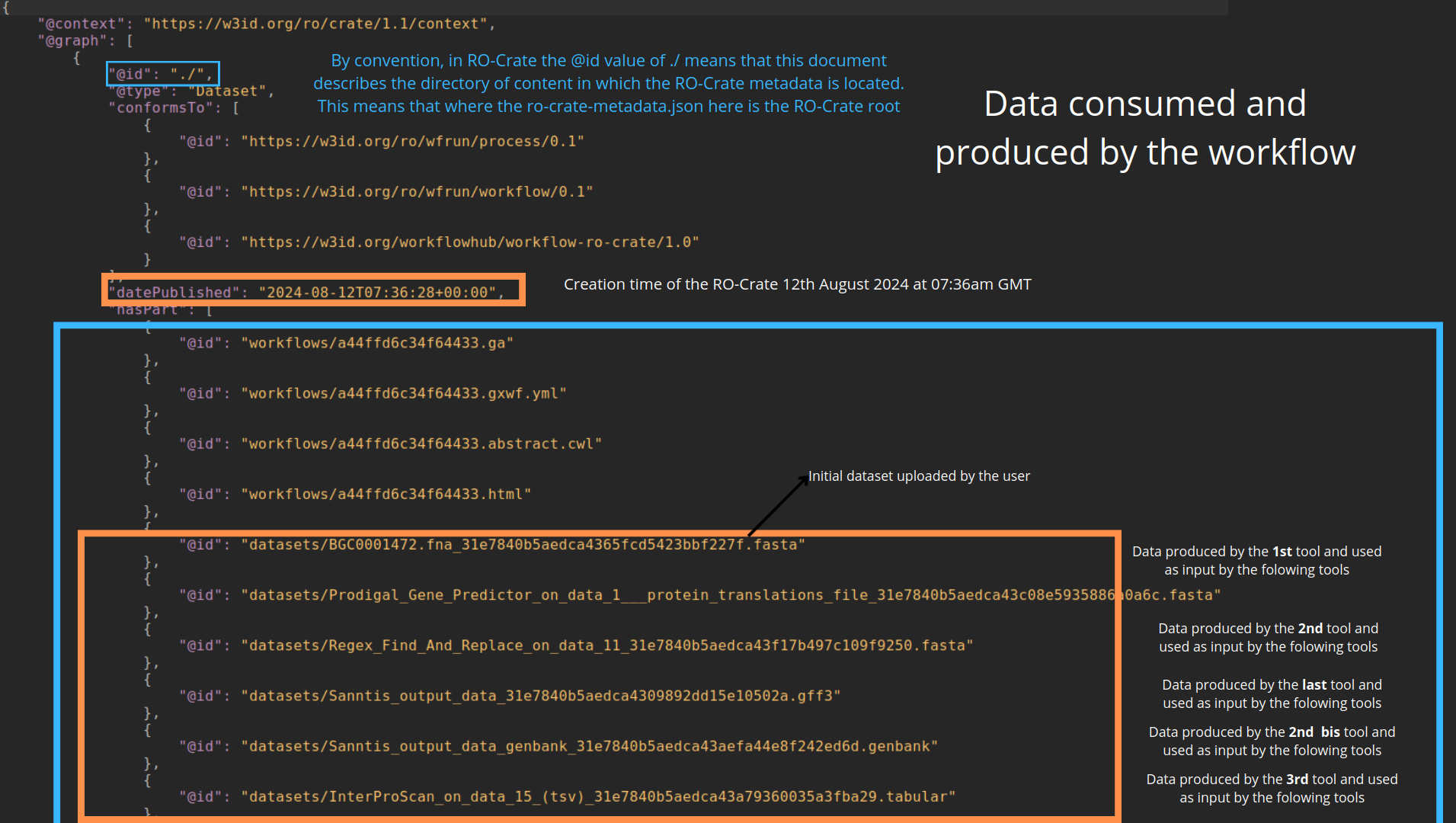 --- ### Description of the RO-Crate structure 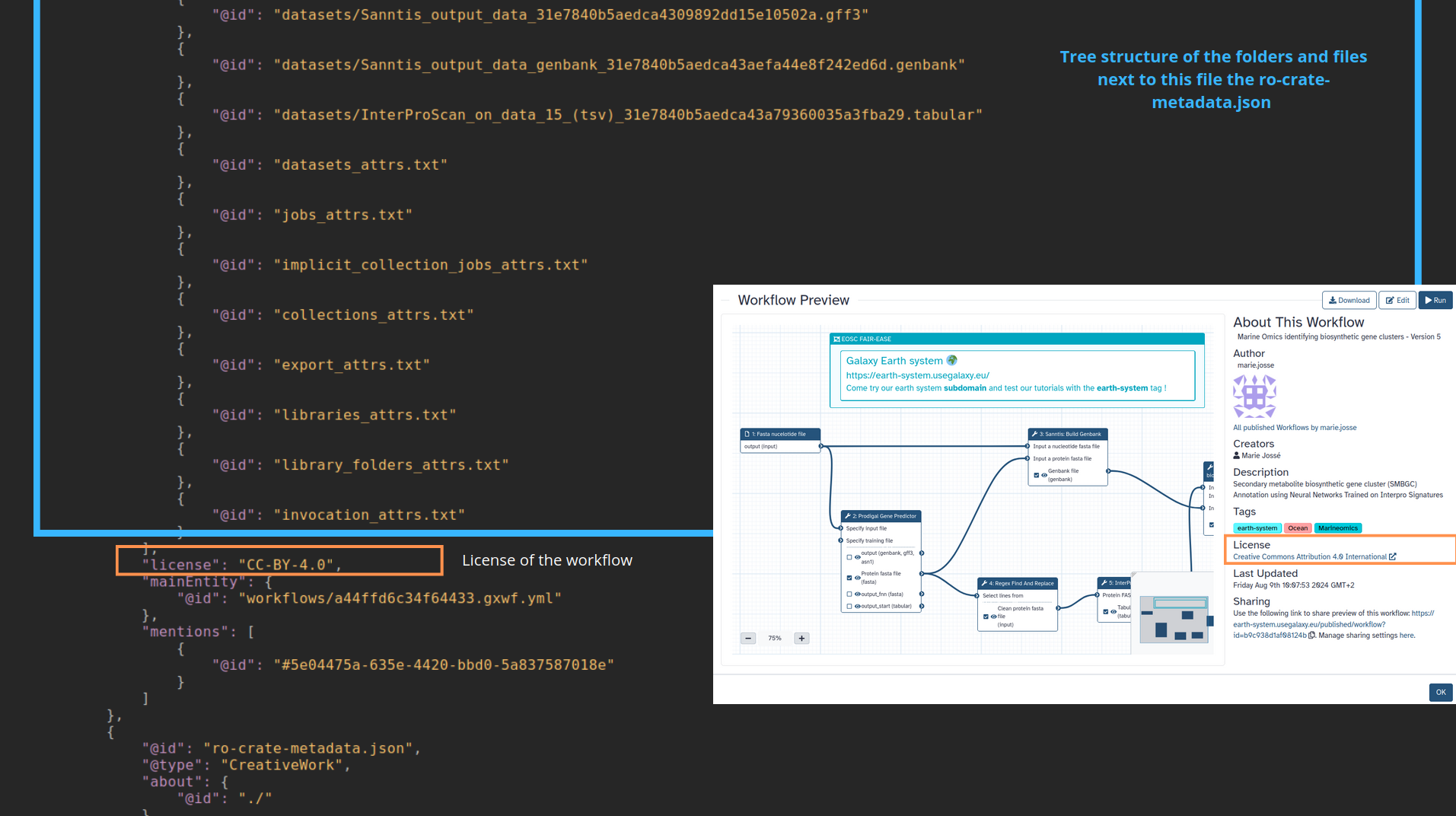 --- ### Galaxy Provenance 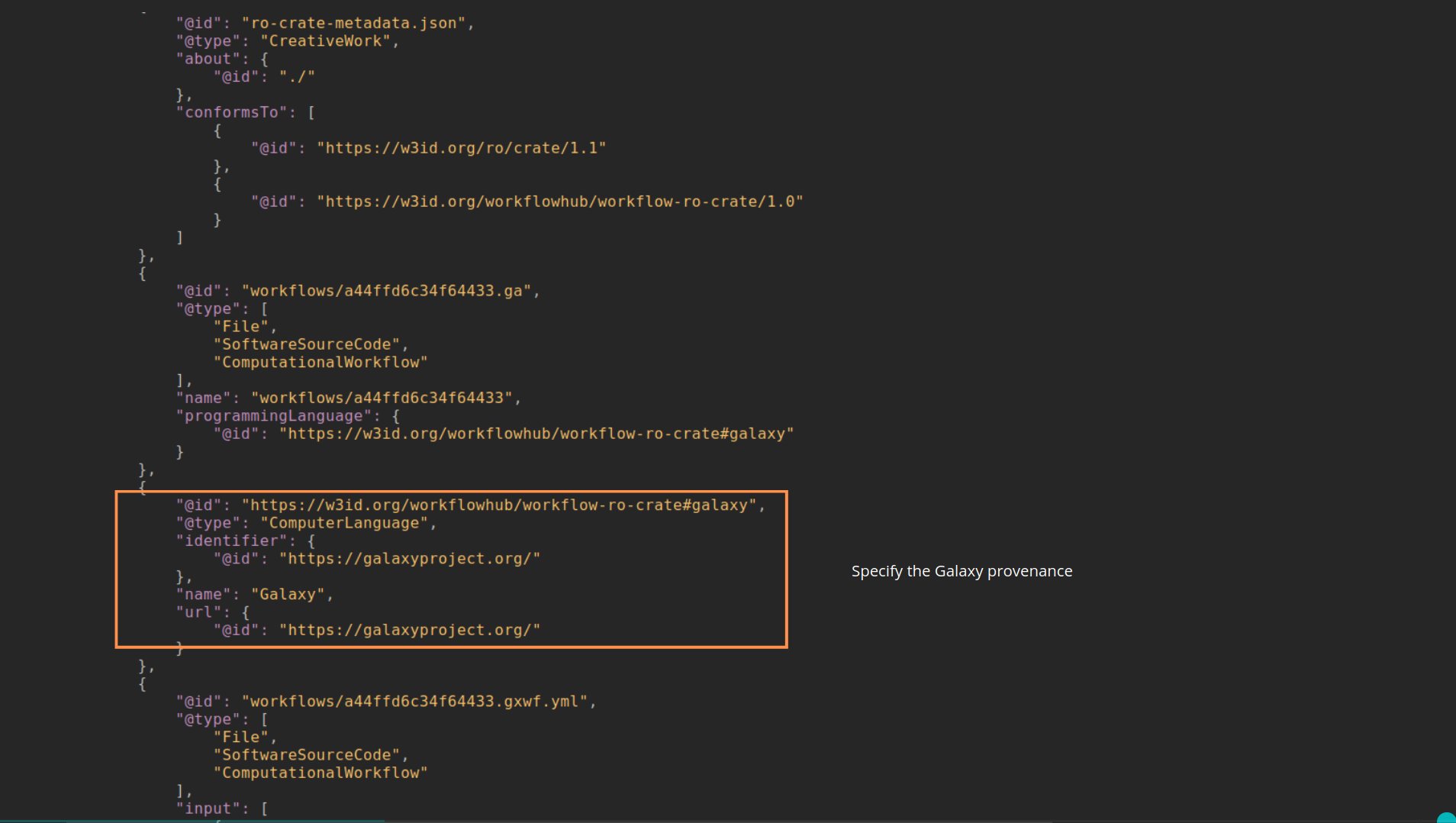 --- ### Title and workflow details 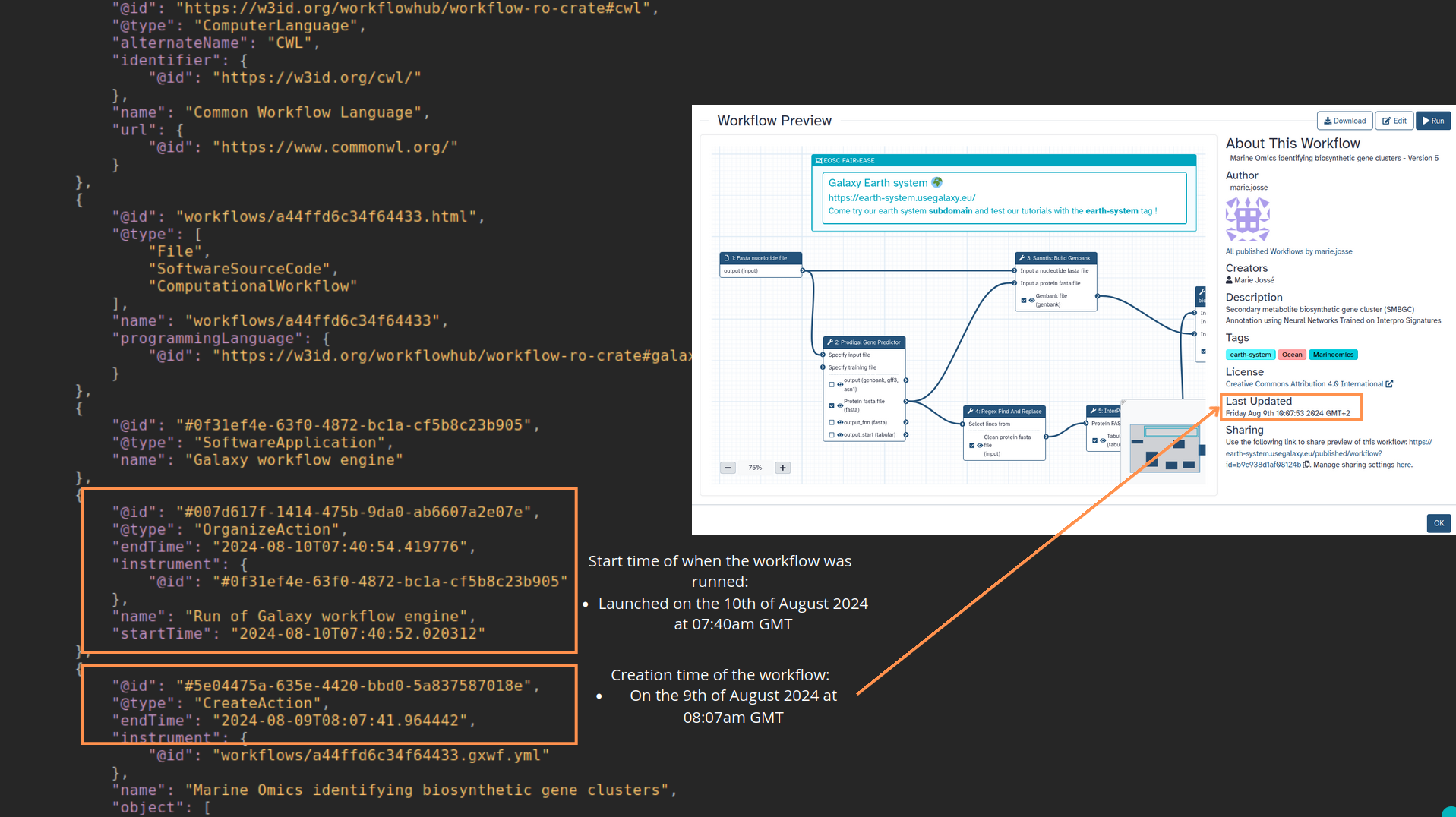 --- ### Dates 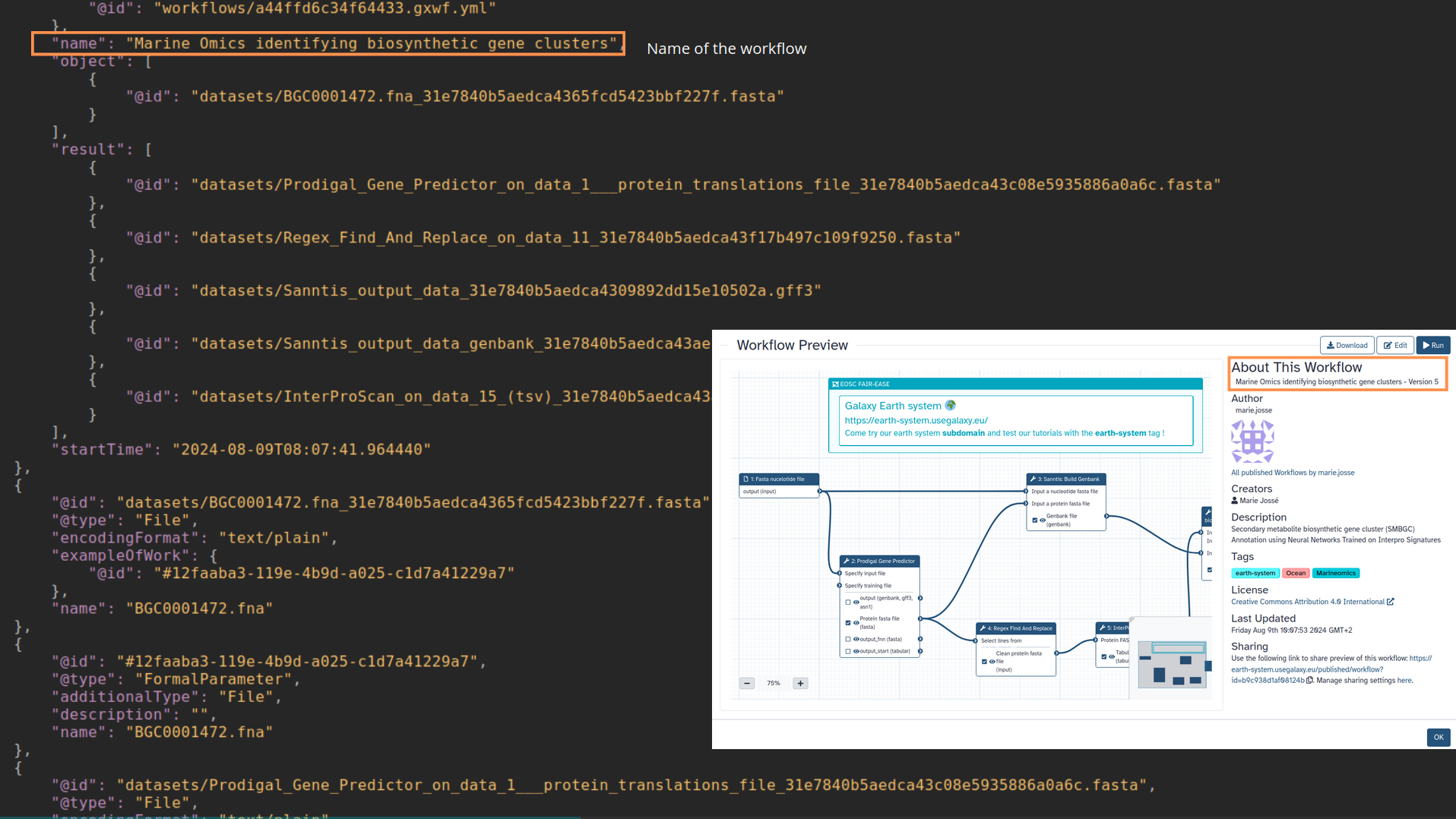 --- ### Data 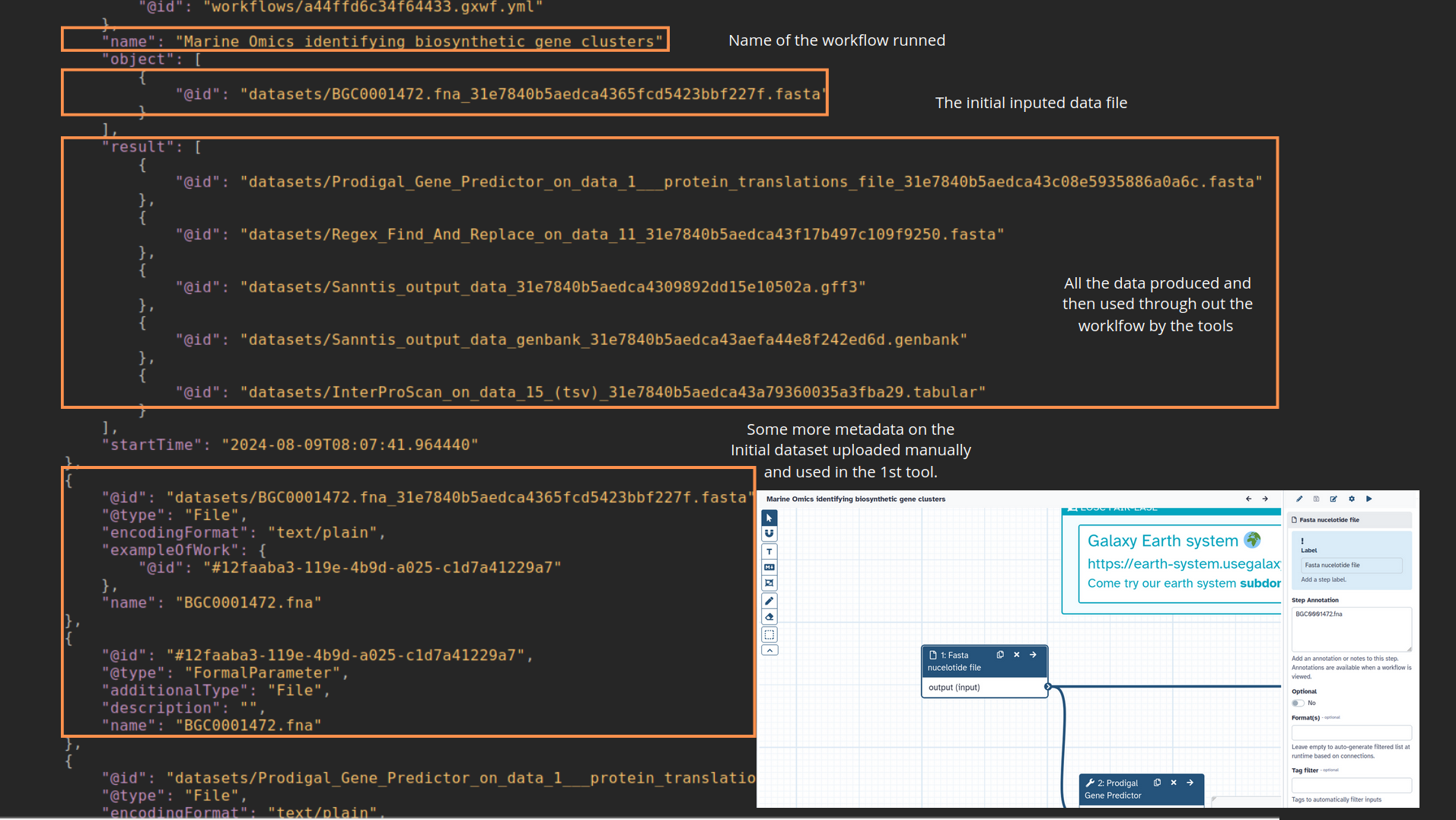 --- ### First tool Prodigal 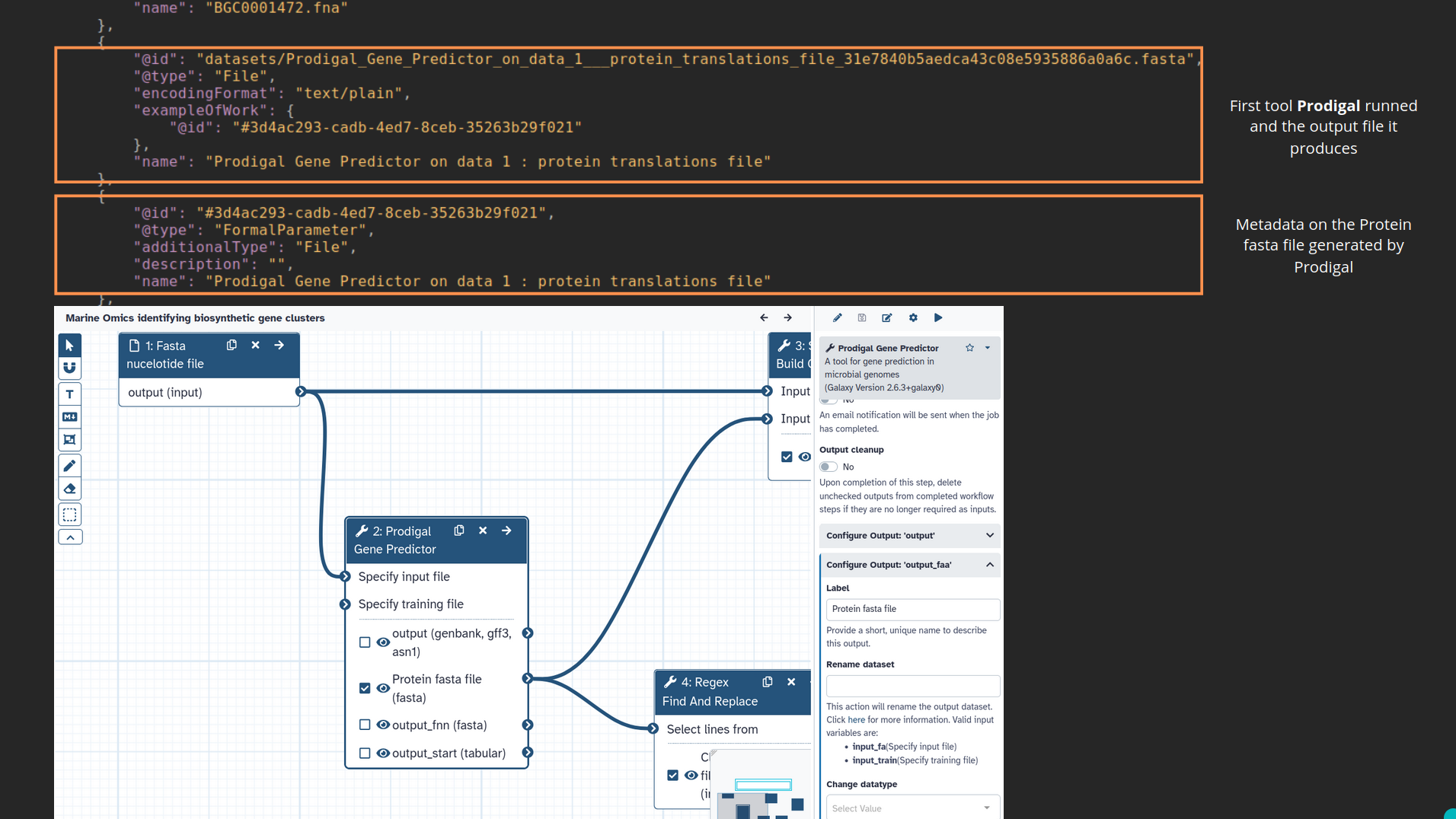 --- ### Second tool Regex Find and Replace 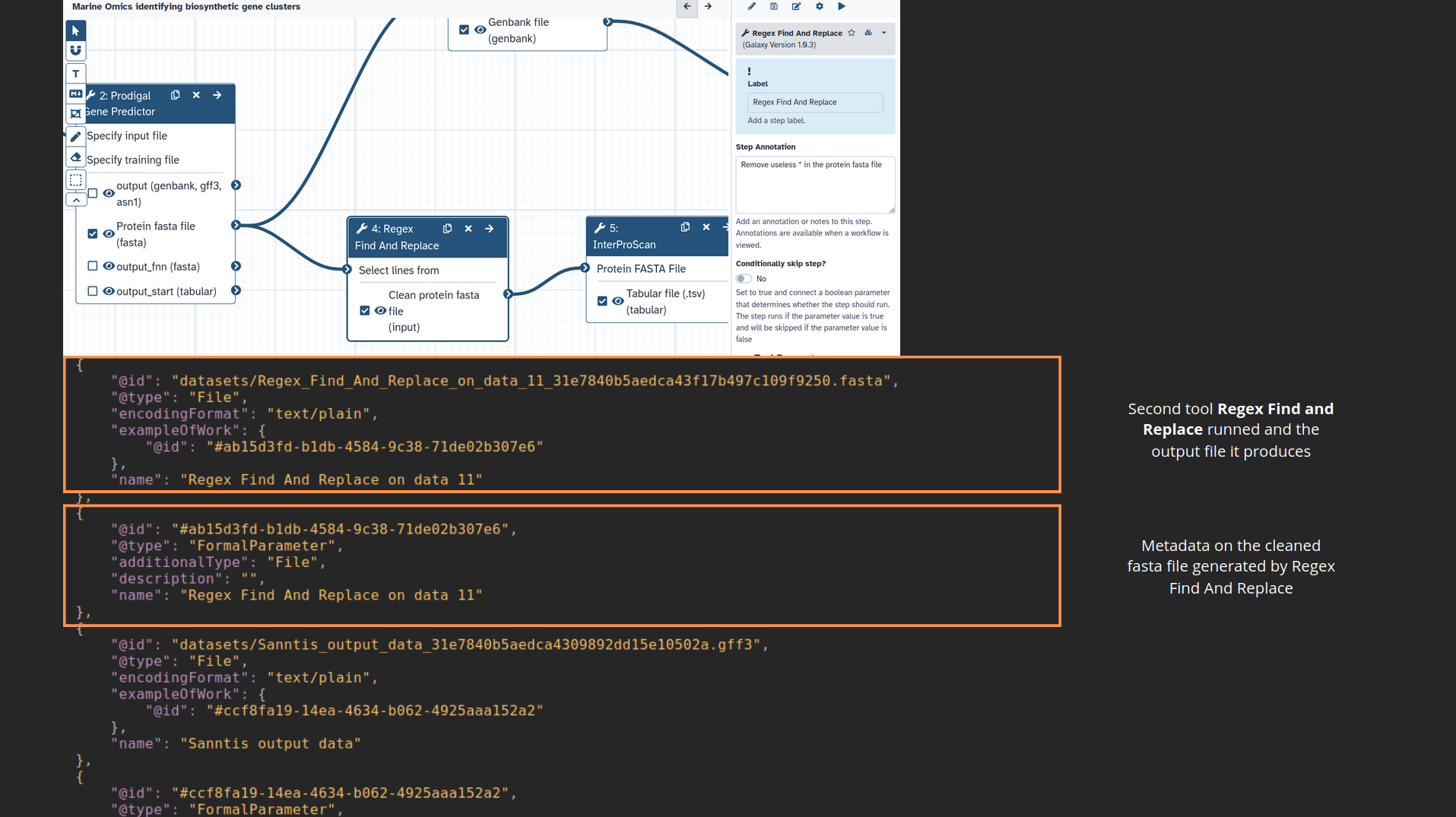 --- ### Third tool InterProScan 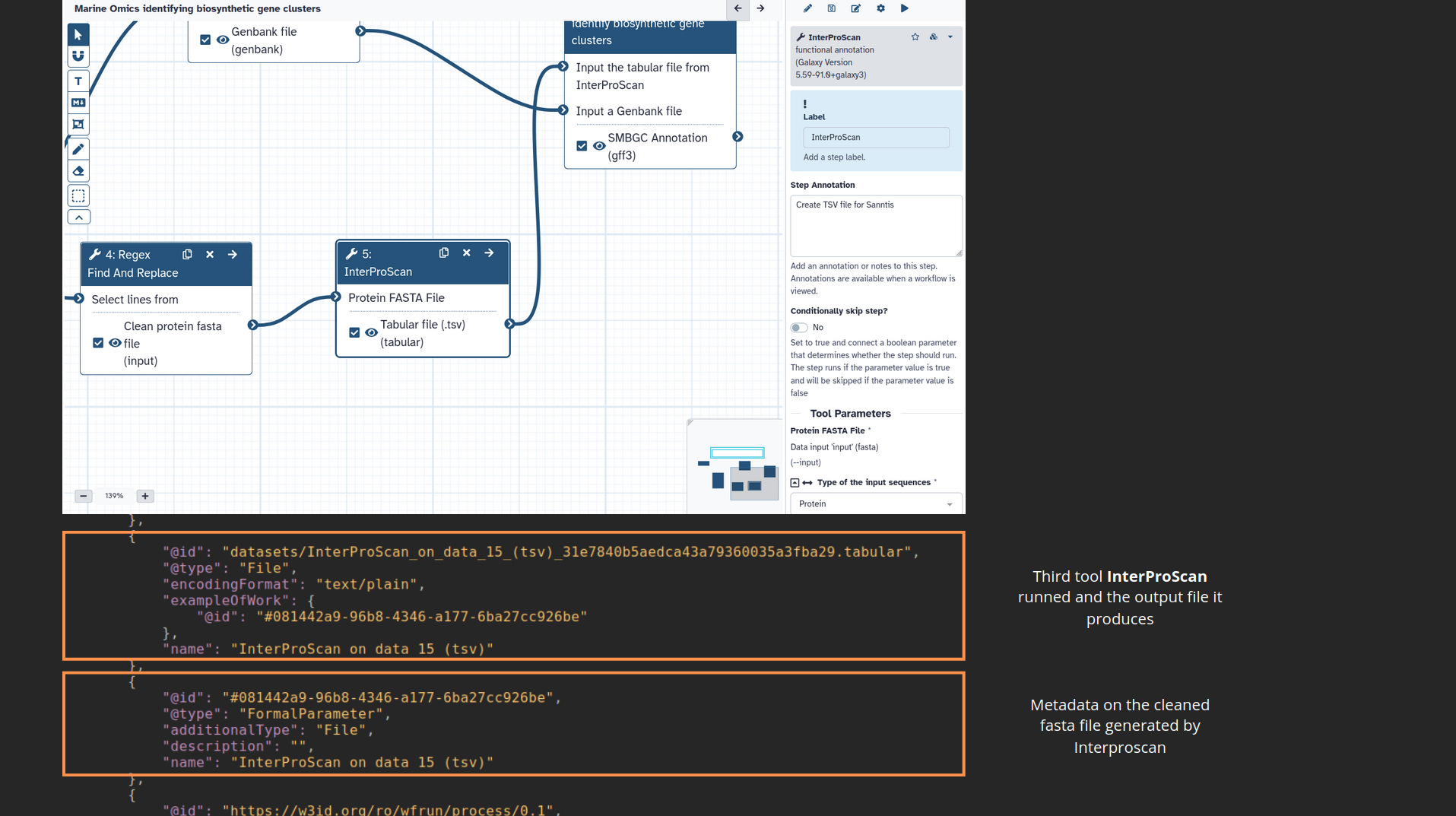 --- ### Fourth tool SanntiS to build Genbank 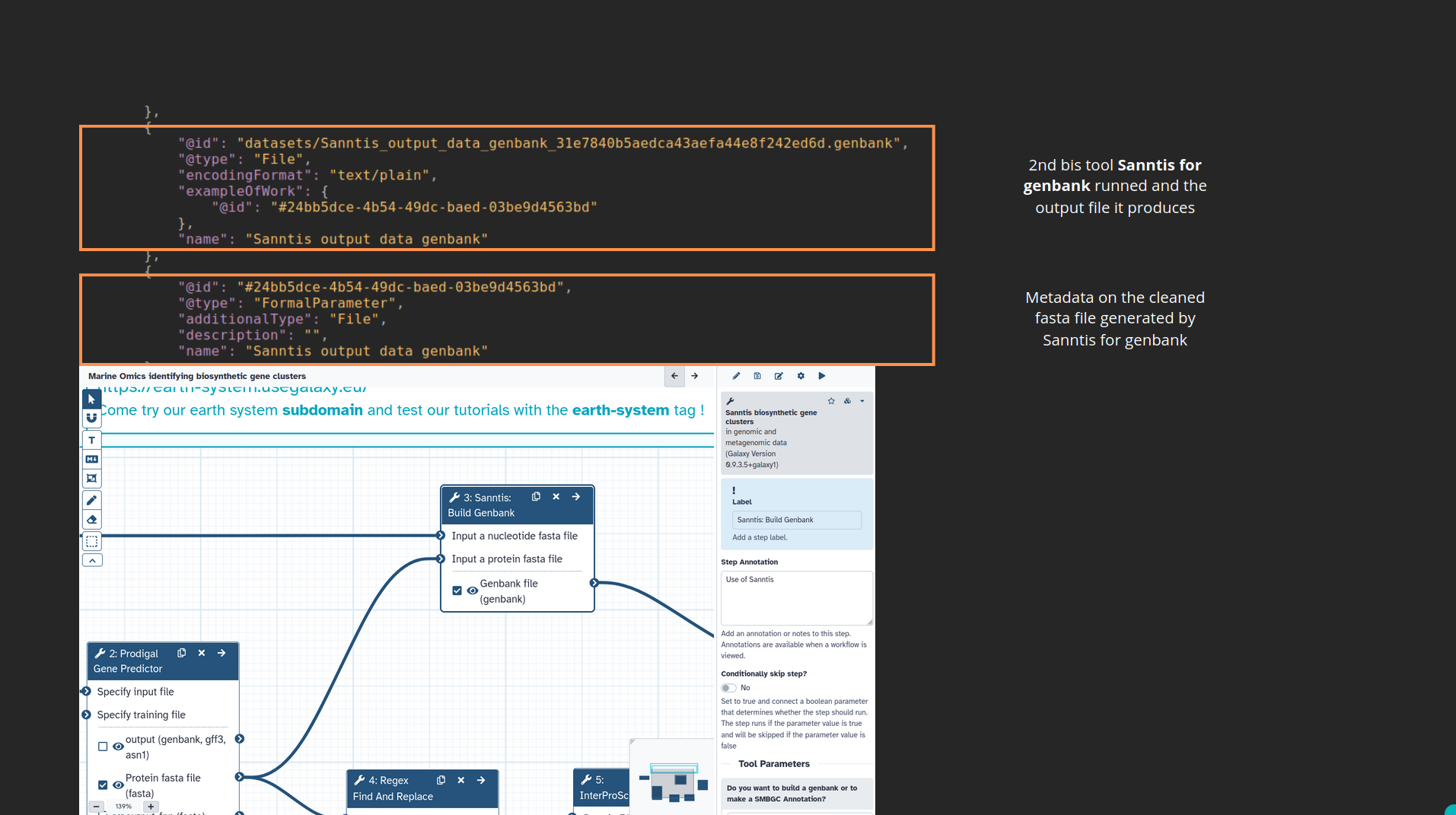 --- ### Fourth tool SanntiS to annotate biosynthetic gene clusters 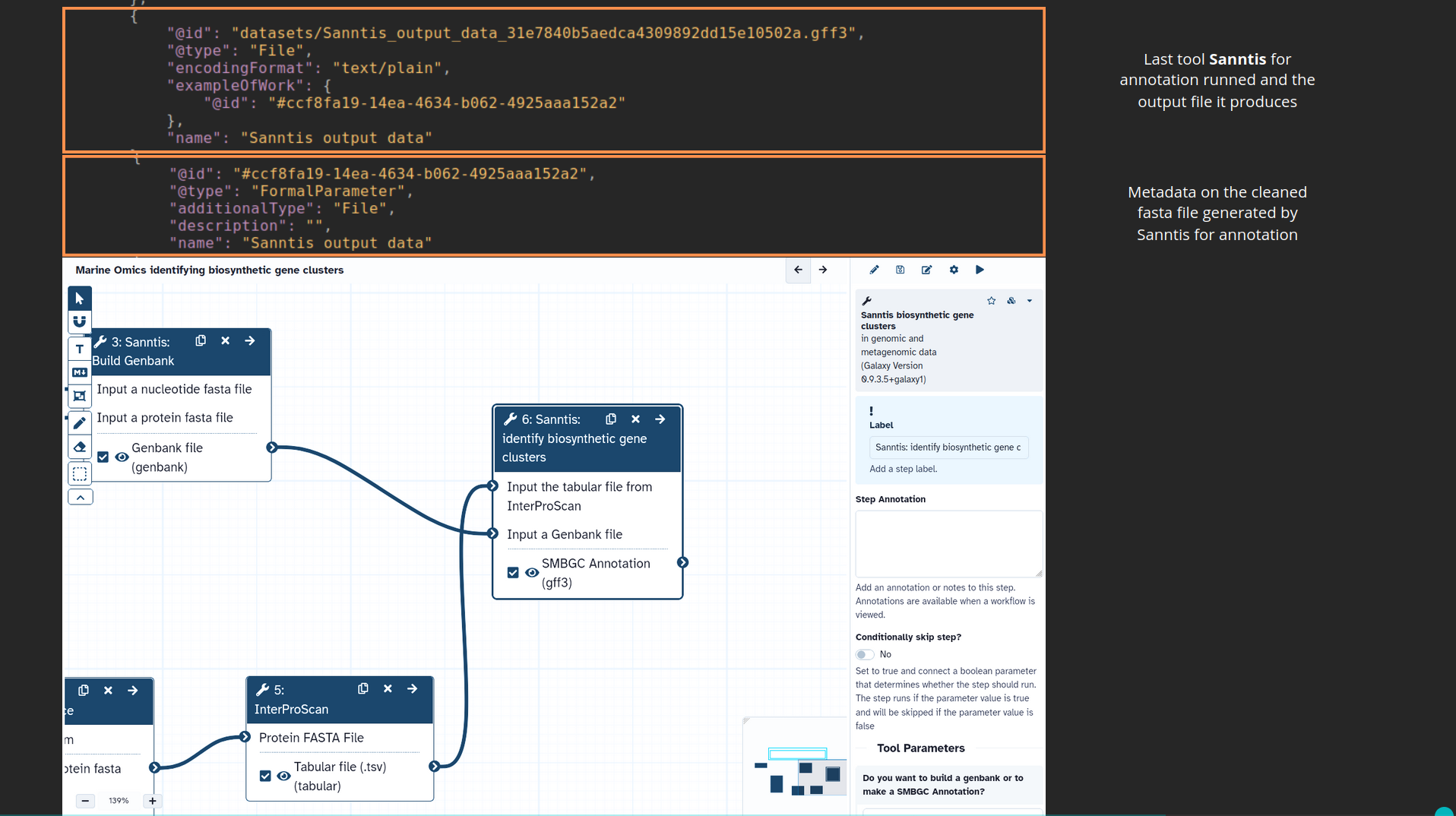 --- ### Details on the different files in the RO-Crate 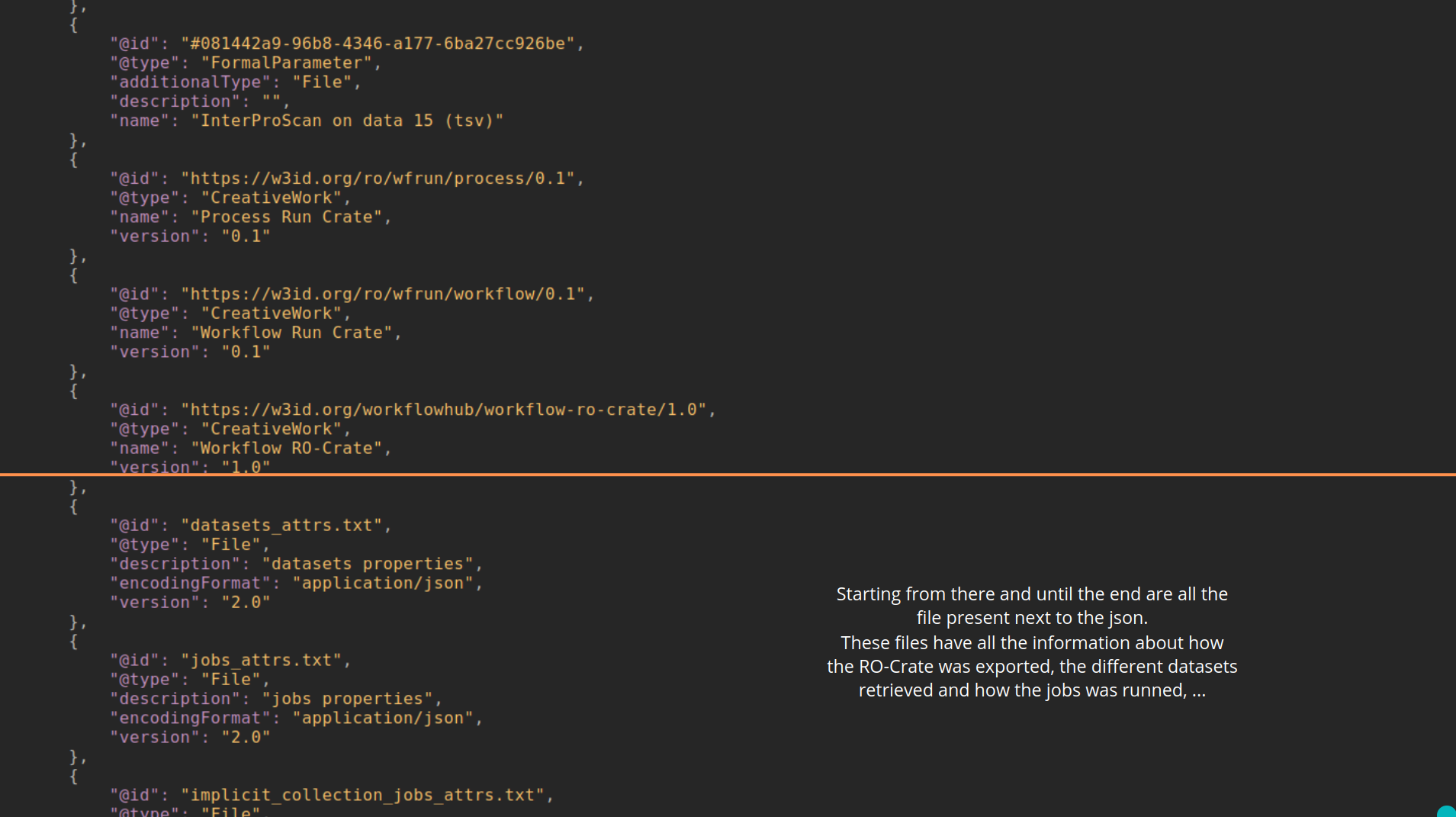 --- ### End of the ro-crate-metadata.json  --- ### Thank you! Try to do the same on your workflow! --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - RO-Crate in earth-system analysis - Marine omics workflow --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/Marie59/" class="contributor-badge contributor-Marie59"><img src="https://avatars.githubusercontent.com/Marie59?s=36" alt="Marie Josse avatar" width="36" class="avatar" /> Marie Josse</a> </td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" style="height: 100px;"/> <div> <div> <img class="funder-avatar" src="https://avatars.githubusercontent.com/fairease" alt="Logo"> </div> <div> </div> </div> <div> <div> <img class="funder-avatar" src="https://avatars.githubusercontent.com/eurosciencegateway" alt="Logo"> </div> <div> </div> </div> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>