name: inverse layout: true class: center, middle, inverse <div class="my-header"><span> <a href="/training-material/topics/sequence-analysis" title="Return to topic page" ><i class="fa fa-level-up" aria-hidden="true"></i></a> <a href="https://github.com/galaxyproject/training-material/edit/main/topics/sequence-analysis/tutorials/mapping/slides.html"><i class="fa fa-pencil" aria-hidden="true"></i></a> </span></div> <div class="my-footer"><span> <img src="/training-material/assets/images/GTN-60px.png" alt="Galaxy Training Network" style="height: 40px;"/> </span></div> --- <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" class="cover-logo"/> <br/> <br/> # Mapping <br/> <br/> <div markdown="0"> <div class="contributors-line"> <ul class="text-list"> <li> <a href="/training-material/hall-of-fame/joachimwolff/" class="contributor-badge contributor-joachimwolff"><img src="https://avatars.githubusercontent.com/joachimwolff?s=36" alt="Joachim Wolff avatar" width="36" class="avatar" /> Joachim Wolff</a> <li> <a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /> Saskia Hiltemann</a> <li> <a href="/training-material/hall-of-fame/EngyNasr/" class="contributor-badge contributor-EngyNasr"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/EngyNasr?s=36" alt="Engy Nasr avatar" width="36" class="avatar" /> Engy Nasr</a> <li> <a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-gallardoalba"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /> Cristóbal Gallardo</a></li> </ul> </div> </div> <!-- modified date --> <div class="footnote" style="bottom: 8em;"> <i class="far fa-calendar" aria-hidden="true"></i><span class="visually-hidden">last_modification</span> Updated: <i class="fas fa-fingerprint" aria-hidden="true"></i><span class="visually-hidden">purl</span><abbr title="Persistent URL">PURL</abbr>: <a href="https://gxy.io/GTN:S00081">gxy.io/GTN:S00081</a> </div> <!-- other slide formats (video and plain-text) --> <div class="footnote" style="bottom: 5em;"> <i class="fas fa-file-alt" aria-hidden="true"></i><span class="visually-hidden">text-document</span><a href="slides-plain.html"> Plain-text slides</a> | <div class="btn-group"> <!-- dropdown with all recordings --> <a href="/training-material/topics/sequence-analysis/tutorials/mapping/recordings/" class="btn btn-default dropdown-toggle topic-icon" data-toggle="dropdown" aria-expanded="false" title="Latest recordings of this material in the GTN Video Library"> <i class="fas fa-video" aria-hidden="true"></i><span class="visually-hidden">video</span> Recordings </a> <ul class="dropdown-menu"> <li><a class="dropdown-item" href="/training-material/topics/sequence-analysis/tutorials/mapping/recordings/index.html#lecture-recording-15-february-2021" title="View the recording for this tutorial"> <i class="fas fa-video" aria-hidden="true"></i><span class="visually-hidden">video</span> Lecture (February 2021) - 10m</a> </li> <li><a class="dropdown-item" href="/training-material/topics/sequence-analysis/tutorials/mapping/recordings/" title="View all recordings for this tutorial"> <i class="fas fa-video" aria-hidden="true"></i><span class="visually-hidden">video</span> View All</a> </li> </ul> </div> </div> <!-- usage tips --> <div class="footnote" style="bottom: 2em;"> <strong>Tip: </strong>press <kbd>P</kbd> to view the presenter notes | <i class="fa fa-arrows" aria-hidden="true"></i><span class="visually-hidden">arrow-keys</span> Use arrow keys to move between slides </div> ??? Presenter notes contain extra information which might be useful if you intend to use these slides for teaching. Press `P` again to switch presenter notes off Press `C` to create a new window where the same presentation will be displayed. This window is linked to the main window. Changing slides on one will cause the slide to change on the other. Useful when presenting. --- ## Requirements Before diving into this slide deck, we recommend you to have a look at: - [Introduction to Galaxy Analyses](/training-material/topics/introduction) --- ### <i class="far fa-question-circle" aria-hidden="true"></i><span class="visually-hidden">question</span> Questions - What is mapping (alignment)? - What is the BAM format? - How can we view aligned sequences? --- ### <i class="fas fa-bullseye" aria-hidden="true"></i><span class="visually-hidden">objectives</span> Objectives - Understand the basic concept of mapping - Learn about factors influencing alignment - See a genome browser used to better understand your aligned data --- # Example NGS pipeline  A high level view of a typical NGS bioinformatics workflow ??? - Mapping step occurs if a reference genome is available for the organism of interest - else: de-novo assembly - Variant calling step is just an example, after mapping can do many steps - Structural Variants / Fusion genes - Differential Gene expression - Alternative Splicing - .. --- # What is mapping? .pull-left[  ] .pull-right[ - Short reads must be combined into longer fragments - **Mapping:** use a reference genome as a guide - **De-novo assembly:** without reference genome ] ??? - Mapping is also referred to as *alignment* - Short reads produced by sequencer must be combined into larger contigs - e.g. reconstruct the chromosomes - mapping uses a reference genome as a guide - can subsequently find where our sample differs from reference (variants) - This tutorial only deals with mapping/alignment - There are other tutorials available for de-novo assembly --- class: top # Sequence alignment - Determine position of short read on the reference genome ``` Reference: . . . A A C G C C T T . . . Read: A G G G G C C T T ``` ??? - Consider situation where we must map this (short) read to this (long) reference - e.g. human genome ~ 3.2 billion base pairs - We scan the reference genome until we find an area that's similar to our read - This area looks pretty similar, but not quite identical.. --- class: top # Sequence alignment - Determine position of short read on the reference genome ``` Reference: . . . A A - C G C C T T . . . | = match . | : - : | | | | | : = mismatch Read: A G G G G C C T T - = gap ``` ??? But if we introduce gaps and allow for some mismatches in bases, this matches up pretty well.. -- - Read could align to multiple places .center[.image-50[]] - How to handle multi-mapped reads? Depends on tool: - Map to best region (but what is "best"? And what about ties?) - Map to all regions - Map to one region randomly - Discard read - How do we determine *best* region? - Assign ***alignment score*** to every mapping ??? Some reads may map to multiple locations - repeat regions, short reads, highly variable regions, sequencing errors, .. We want a way to determine *best* alignment if none are perfect matches.. --- class: top # Alignment Scoring (basics) - **Reward** for a match (e.g. +10), **penalty** for a mismatch (e.g. -5) - **Penalty** for gaps - *Linear:* every gap same penalty (e.g. -5) - *Affine:* gap open vs gap extend (e.g. -5 and -1) - Different tools use different scoring values (and give different results) .center[ .image-25[] **Example** (with affine gap penalty) ] ??? - Each locus get scored independently (first row of scores in example) - Scores from all loci are added up (cumulative score row) - Final score for entire alignment in this example is 19 - These reward and penalty values are just examples and will vary --- class: top # Alignment Scoring (advanced) - **Base quality** - Mismatch of low-confidence base: lower penalty - Mismatch of high-confidence base: higher penalty - **Transitions vs transversions** - Transitions about 2x as frequent as transversions .center[ .image-50[  ] .image-25[  ] ] - Knowledge about sequencing platform and biases - Optimize for read length, error rate, homopolymer accuracy, etc.. .footnote[More information about mapping algorithms: [10.1089/cmb.2012.0022](https://doi.org/10.1089/cmb.2012.0022)] ??? Many more complexities may be considered, different tools make different choices Transitions are more likely to occur in real sequences, so may give lower penalty than transversions **Transitions** are interchanges of two-ring purines (A G) or of one-ring pyrimidines (C T): they therefore involve bases of similar shape. **Transversions** are interchanges of purine for pyrimidine bases, which therefore involve exchange of one-ring and two-ring structures.  --- # Looks easy but.. --- class: top # Sequence Alignment ``` Reference: AAA CAGTGA GAA Observed: AAA TCTCT GAA ``` ??? Suppose we want to map this read (bottom) to this reference sequence (top) --- class: top # Sequence Alignment ``` Reference: AAA CAGTGA GAA Observed: AAA TCTCT GAA ``` <table style="width:100%; table-layout: fixed; font-size:0.8em"> <th>Alignment</th><th></th> <tr><td><pre> AAA-CAGTGAGAA |||-|--|::||| AAATC--TCTGAA </pre></td> <td>Maybe like this?</td> </tr> </table> ??? This is one possibility, is it the only one? --- class: top # Sequence Alignment ``` Reference: AAA CAGTGA GAA Observed: AAA TCTCT GAA ``` <table style="width:100%; table-layout:fixed; font-size:0.8em;"> <th>Alignment</th><th></th> <tr><td><pre> AAA-CAGTGAGAA |||-|--|::||| AAATC--TCTGAA </pre></td> <td>Maybe like this?</td> </tr> <tr><td><pre> AAACAGTGAGAA |||-::|::||| AAA-TCTCTGAA </pre></td> <td> Or this? </td> </tr> </table> ??? This is also a possible alignment. Not easy to say which is better. --- class: top # Sequence Alignment ``` Reference: AAA CAGTGA GAA Observed: AAA TCTCT GAA ``` <table style="width:100%; table-layout:fixed; font-size:0.8em"> <th>Alignment</th><th></th> <tr><td><pre> AAA-CAGTGAGAA |||-|--|::||| AAATC--TCTGAA </pre></td> <td>Maybe like this?</td> </tr> <tr><td><pre> AAACAGTGAGAA |||-::|::||| AAA-TCTCTGAA </pre></td> <td> Or this? </td> </tr> <tr><td><pre> AAACAGTGAGAA |||:-:|::||| AAAT-CTCTGAA </pre></td> <td>Or..? </td> </tr> </table> ??? And a third option --- class: top # Sequence Alignment ``` Reference: AAA CAGTGA GAA Observed: AAA TCTCT GAA ``` <table style="width:100%; table-layout:fixed; font-size:0.8em"> <th>Alignment</th><th></th> <tr><td><pre> AAA-CAGTGAGAA |||-|--|::||| AAATC--TCTGAA </pre></td> <td>Maybe like this?</td> </tr> <tr><td><pre> AAACAGTGAGAA |||-::|::||| AAA-TCTCTGAA </pre></td> <td> Or this? </td> </tr> <tr><td><pre> AAACAGTGAGAA |||:-:|::||| AAAT-CTCTGAA </pre></td> <td>Or..? </td> </tr> <tr><td><pre> AAACAGTCA-----GAA |||-----------||| AAA------TCTCTGAA </pre></td> <td> What about this? </td> </tr> </table> ??? There is no one right way to do alignment - Hard to say which of these is "better" or "worse" - Just different choices, but all valid Mapping is a non-trivial problem! --- class: top # Sequence Alignment ``` Reference: AAA CAGTGA GAA Observed: AAA TCTCT GAA ``` <table style="width:100%; table-layout:fixed; font-size:0.8em"> <th>Alignment</th><th>Tool</th> <tr><td><pre> AAA-CAGTGAGAA |||-|--|::||| AAATC--TCTGAA </pre></td> <td>Novoalign</td> </tr> <tr><td><pre> AAACAGTGAGAA |||-::|::||| AAA-TCTCTGAA </pre></td> <td> Ssaha2 </td> </tr> <tr><td><pre> AAACAGTGAGAA |||:-:|::||| AAAT-CTCTGAA </pre></td> <td> BWA </td> </tr> <tr><td><pre> AAACAGTCA-----GAA |||-----------||| AAA------TCTCTGAA </pre></td> <td> Complete Genomics </td> </tr> </table> ??? We didn't just make these up, these real aligners gave these different results --- class: top # Sequence Alignment ``` Reference: AAA CAGTGA GAA Observed: AAA TCTCT GAA ``` <table style="width:100%; table-layout:fixed; font-size:0.8em"> <th>Alignment</th><th>Variant calls</th> <tr><td><pre> AAA-CAGTGAGAA |||-|--|::||| AAATC--TCTGAA </pre></td> <td><pre> ins T del AG sub GA -> CT </pre></td> </tr> <tr><td><pre> AAACAGTGAGAA |||-::|::||| AAA-TCTCTGAA </pre></td> <td><pre> del C sub AG -> TC sub GA -> CT </pre></td> </tr> <tr><td><pre> AAACAGTGAGAA |||:-:|::||| AAAT-CTCTGAA </pre></td> <td><pre> snp C -> T del A snp G -> C sub GA -> CT </pre></td> </tr> <tr><td><pre> AAACAGTGA-----GAA |||-----------||| AAA------TCTCTGAA </pre></td> <td><pre> del CAGTGA ins TCTCT </pre></td> </tr> </table> ??? **Important:** Mapping can affect downstream analysis! These different mappings led to different variants, and hard to tell they are equivalent. --- # Try it yourself! - Lego time! Who wants to volunteer? - Or try this [online sequence alignment game](http://web.archive.org/web/20200411075748/https://teacheng.illinois.edu/SequenceAlignment/): <!-- using webarchive version because game seems broken, once fixed we can update the link back to: http://teacheng.illinois.edu/SequenceAlignment/ --> .image-75[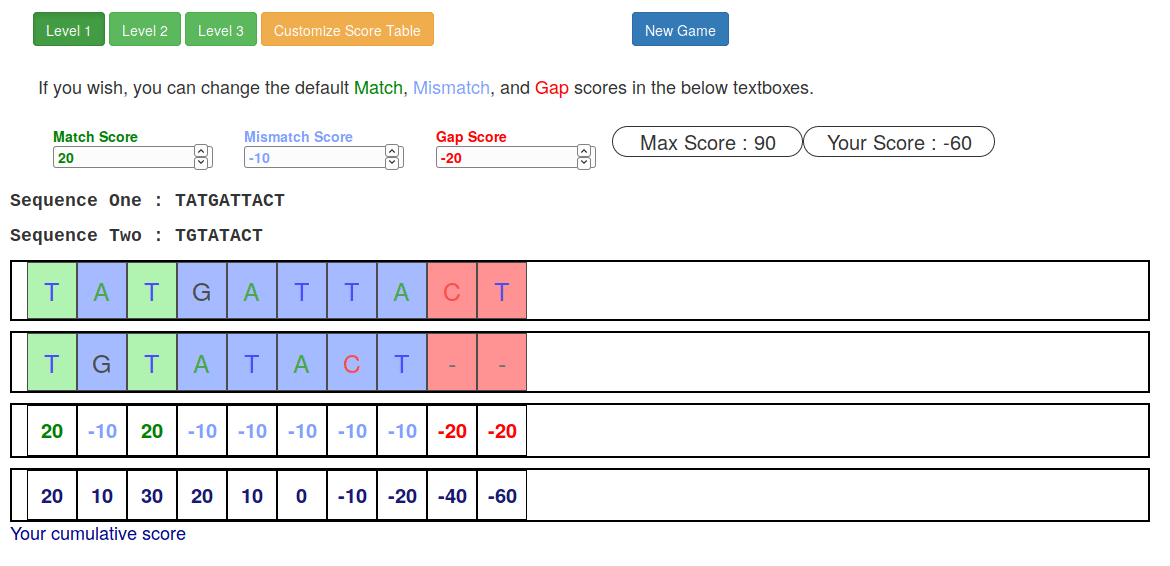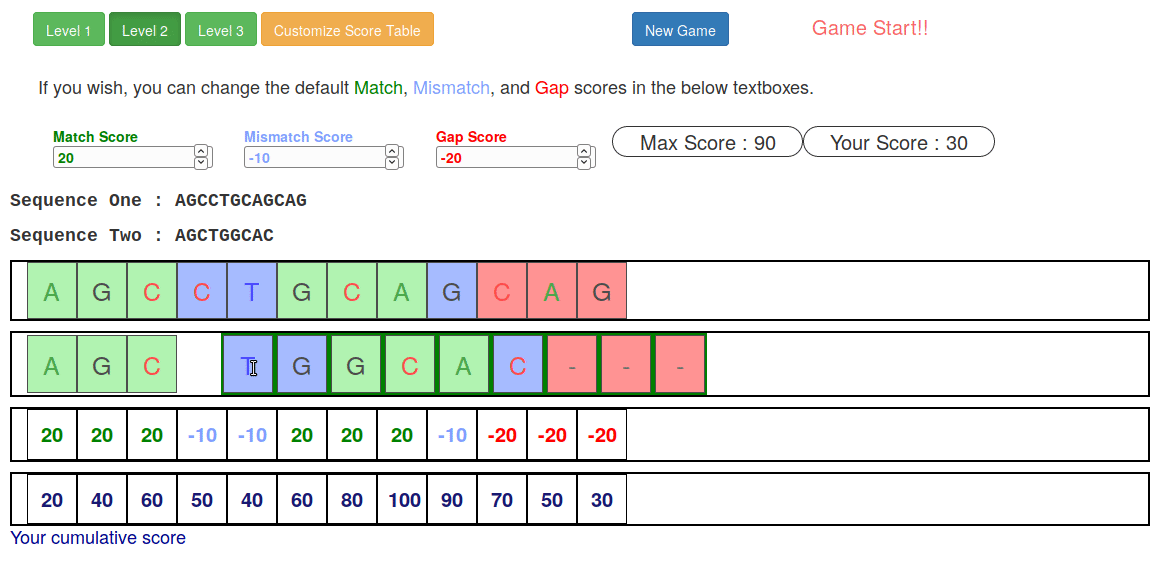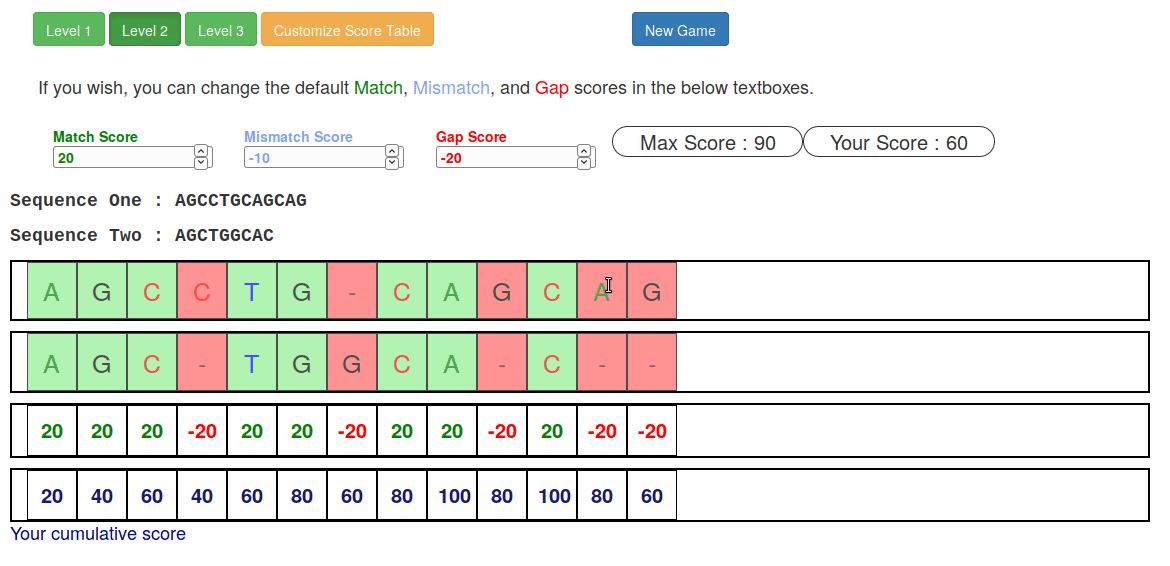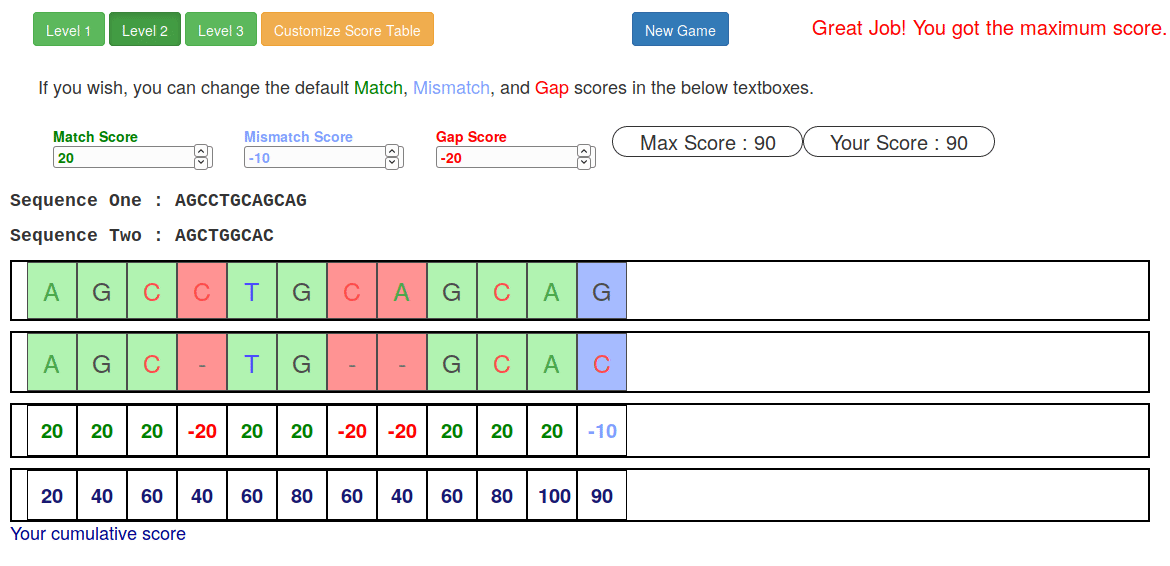] .footnote[https://tinyurl.com/sequence-alignment] ??? Can have learners play around with this alignment game now Or use Lego bricks, each nucleotide a different colour --- ## Paired-end sequencing - **Sequencing:** Cut longer fragments of DNA, sequence only the ends .center[.image-90[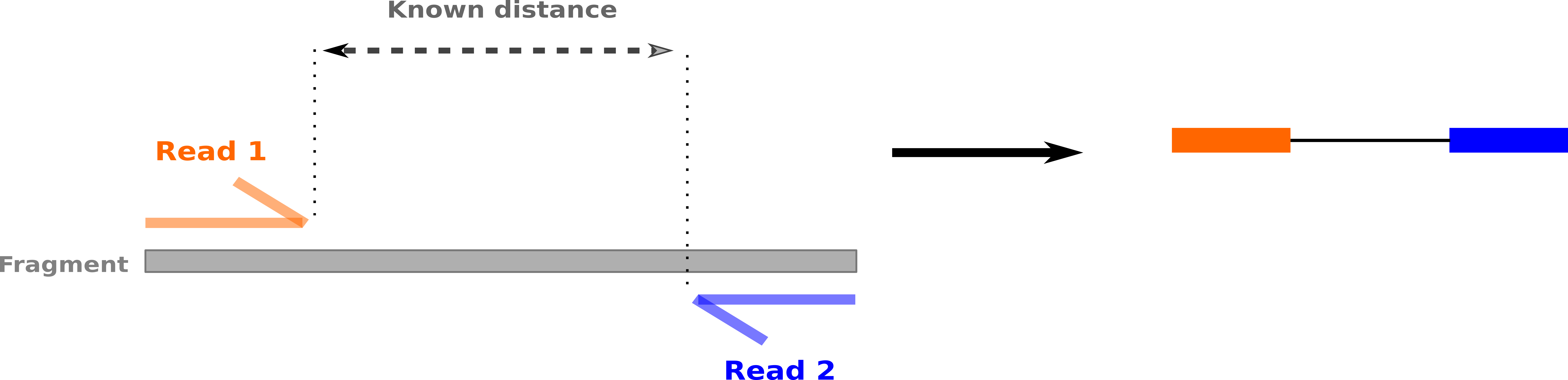]] - **Mapping:** known distance between reads improves accuracy .center[.image-75[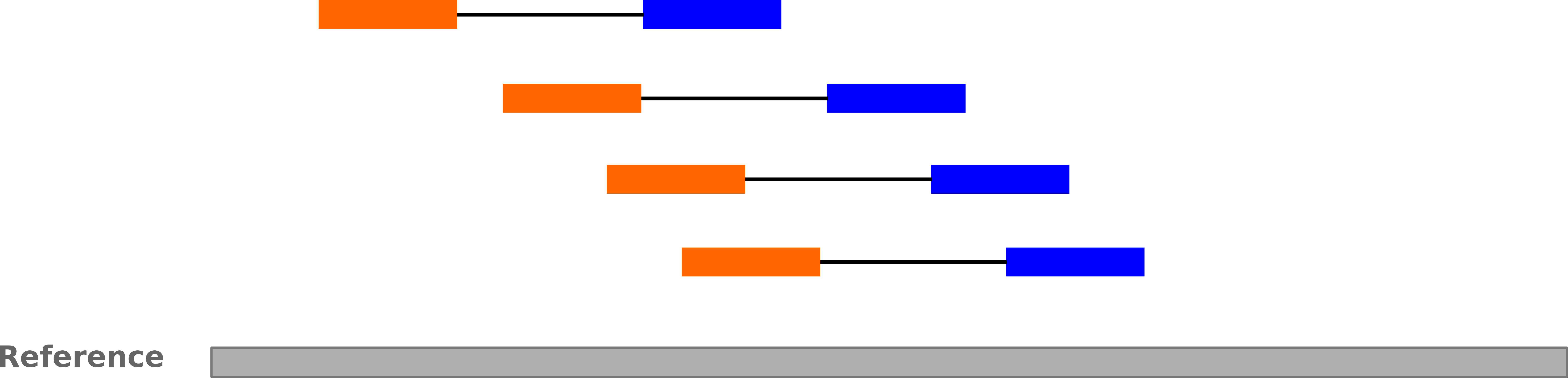]] ??? - The fragments are too long to sequence entirely, but we can sequence the ends. - Then we have the added information of how far apart these two reads must map - This improves our mapping - For example for multi-mapped reads, or repeats (next slide) --- class: top ## Repeats - Multi-mapped reads (e.g. because of repeats) may now be resolved - **Single-end:**  ??? In the case of repeats, a single-end read alone would not have be enough for unique mapping.. -- - **Paired-end:** 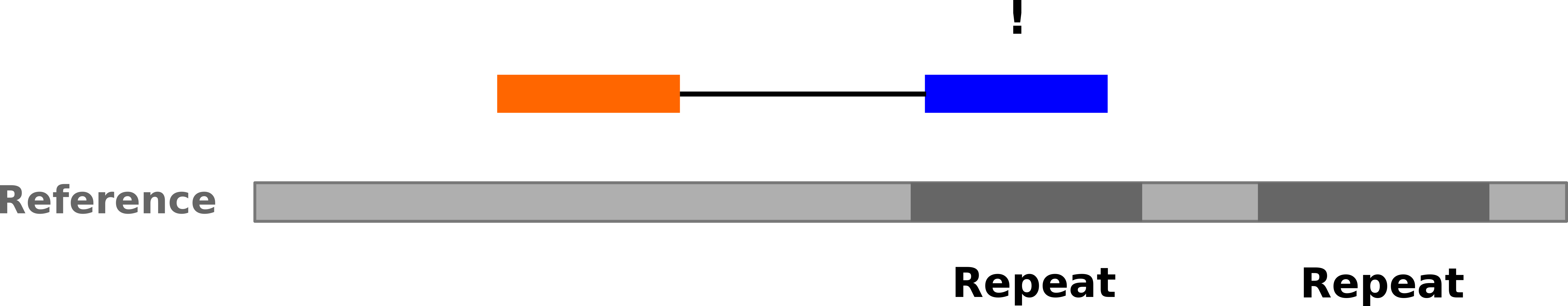 ??? But with the additional information provided by paired-end protocol (distance to mate), this can now be resolved.. --- class: top # InDels (Insertions / Deletions) - Discordant insert size may indicate insertion or deletion between reads - **Deletions:** Longer mapping distance than expected .image-75[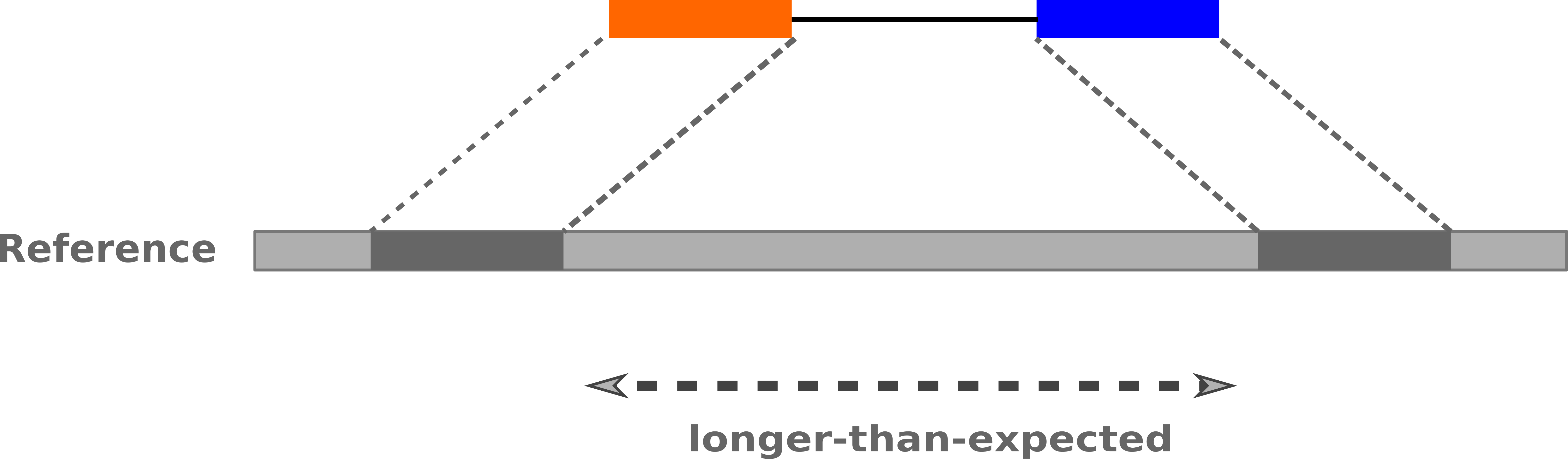] -- - **Insertions:** Shorter mapping distance than expected .image-75[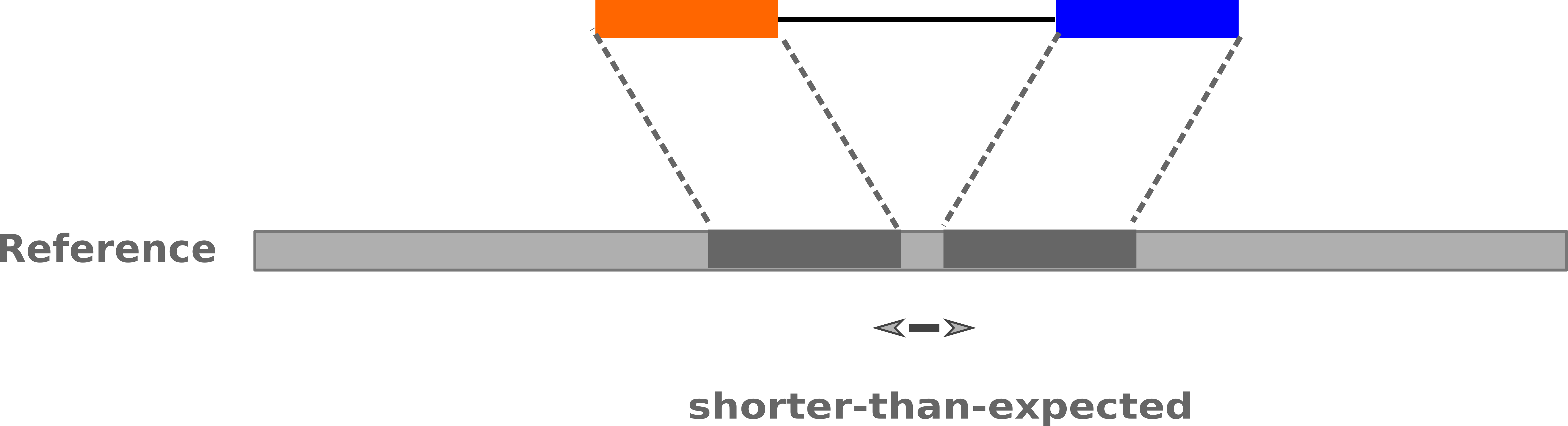] ??? - Unexpected mapping distance between two reads in a pair may indicate a variant. - Exact location of variant unknown unless more reads covering the area - Only know it it somewhere between the two reads **FAQ:** "What about mate-pair sequencing?" - Same concept as paired-end - Much longer distance between ends - Very different library prep - Useful for detection of larger Structural Variations (SVs) / Fusion Genes - longer than expected distance between mates: deletion in sample - shorter than expected distance beetween mates: insertion in sample - unexpected orientation of one mate: inversion in sample --- class: top ## Paired-end FASTQ files - Sequencer produces two FASTQ files: - **Forward** reads (usually **`_1`** or **`_R1`** in file name) - **Reverse** reads (usually **`_2`** or **`_R2`** in file name)  ??? When you have paired-end data, you will usually get 2 files. - File names identical except for e.g. `_1`/`_2` or `_R1`/`_R2` - First file contains all the forward reads ("left" sides of pairs) - Other file contains all the reverse reads Pairing also visible in read names - `/1` `/2` at end or `1:` and `2:` in read ID -- - Sometimes: One **interleaved** (or **interlaced**) FASTQ file - Most tools require 2 separate files - <i class="fas fa-wrench" aria-hidden="true"></i><span class="visually-hidden">tool</span> De-interlace tools in Galaxy for conversion ??? Sometimes data can be in a single **interleaved file** (aka **interlaced**) - alternating forward and reverse read - de-interlace tools in Galaxy to convert this to two separate files - because many tools require two separate files --- class: top ## Paired-end FASTQ files - Order of reads matters! - **`N`<sup>th</sup>** read in forward file <i class="fa fa-arrows-h" aria-hidden="true"></i> **`N`<sup>th</sup>** read in reverse file - Much faster than determining pairing by read names alone - ***Always trim and filter together!*** ??? Most tools blindly assume that first read in forward file is paired with first read in reverse file etc Otherwise too slow - for every read, worst case have to scan all reads in other file - for files with millions of reads, that is millions ^ millions When trimming and filtering, if a read is removed from one file, its mate must be removed from other one too! **Always trim together in paired-end mode!** -- .pull-left[ .red[ ``` @PAIR-1 forward GGGTGATGGCCGCTGCCGATGGCGTCAAAT + ))%255CCF>>>>>>CCCCCCC65`IIII% ``` ] .orange[ ``` @PAIR-2 forward GATTTGGGGTTCAAAGCAGTATCGATCAA + !''3((((^^d+))%%%++)(%%%%).1) ``` ] .blue[ ``` @PAIR-3 forward TCGCACTCAACGCCCTGCATATGACAAGAC + A64;##=#B9=AAAAAAAAAA9#:AB95%^ ``` ] **`mysample_R1.fastq`** ] .pull-right[ <i class="fa fa-arrows-h" style="position:absolute;font-size:3em;left:8em;"></i> .red[ ``` @PAIR-1 reverse AAGTTACCCTTAACAACTTAAGGGTTTTCA + fffddf`feedB`IABa)^%YBBBRTT\^d ``` ] <i class="fa fa-arrows-h" style="position:absolute;font-size:3em;left:8em;"></i> .orange[ ``` @PAIR-2 reverse AGCAGAAGTCGATGATAATACGCGTCGTTT + IIIIIII^^IIId`?III%IIIGII>IIII ``` ]<i class="fa fa-arrows-h" style="position:absolute;font-size:3em;left:8em;"></i> .blue[ ``` @PAIR-3 reverse AATCCATTTGTTCAACTCACAGTTTACCGT + 9C;=;=<9@4868>9:67AA<9>65<=>59 ``` ] **`mysample_R2.fastq`** ] ??? - Nth read in forward file belongs in a pair with Nth read in reverse file - So red reads in this slide form a pair, orange ones, etc --- class: top ## Paired-end FASTQ files - Order of reads matters! - **`N`<sup>th</sup>** read in forward file <i class="fa fa-arrows-h" aria-hidden="true"></i> **`N`<sup>th</sup>** read in reverse file - Much faster than determining pairing by read names alone - ***Always trim and filter together!*** .pull-left[ <i class="fa fa-arrows-h" style="position:absolute;font-size:3em;left:8em;"></i> .red[ ``` @PAIR-1 forward GGGTGATGGCCGCTGCCGATGGCGTCAAAT + ))%255CCF>>>>>>CCCCCCC65`IIII% ``` ] .left[<i class="fa fa-cut" style="width:15%;position:absolute;font-size:5em;"></i>] <i class="fa fa-arrows-h" style="position:absolute;font-size:3em;left:8em;"></i> .orange[ ``` @PAIR-2 forward GATTTGGGGTTCAAAGCAGTATCGATCAA + !''3((((^^d+))%%%++)(%%%%).1) ``` ] <i class="fa fa-arrows-h" style="position:absolute;font-size:3em;left:8em;"></i> .blue[ ``` @PAIR-3 forward TCGCACTCAACGCCCTGCATATGACAAGAC + A64;##=#B9=AAAAAAAAAA9#:AB95%^ ``` ] **`mysample_R1.fastq`** ] .pull-right[ .red[ ``` @PAIR-1 reverse AAGTTACCCTTAACAACTTAAGGGTTTTCA + fffddf`feedB`IABa)^%YBBBRTT\^d ``` ] .orange[ ``` @PAIR-2 reverse AGCAGAAGTCGATGATAATACGCGTCGTTT + IIIIIII^^IIId`?III%IIIGII>IIII ``` ] .blue[ ``` @PAIR-3 reverse AATCCATTTGTTCAACTCACAGTTTACCGT + 9C;=;=<9@4868>9:67AA<9>65<=>59 ``` ] **`mysample_R2.fastq`** ] ??? - Important to always provide both files to trimming and filtering tools together - If a read in one file gets removed (e.g. because it is below quality threshold), but it's mate is not, the pairing between the two files is no longer correct. - If one half of pair is trimmed, the other - also removed, or - put into separate "singletons" FASTQ file that some mappers can use - FAQ:" why not look at read names to determine pairing?" - analysis would be much slower if for every read must scan (max) entire other file for mate, since often millions or reads (for whole-genome sequencing). --- class: top ## Paired-end FASTQ files - Order of reads matters! - **`N`<sup>th</sup>** read in forward file <i class="fa fa-arrows-h" aria-hidden="true"></i> **`N`<sup>th</sup>** read in reverse file - Much faster than determining pairing by read names alone - ***Always trim and filter together!*** .pull-left[ <i class="fa fa-arrows-h" style="position:absolute;font-size:3em;left:8em;"></i> .red[ ``` @PAIR-1 forward GGGTGATGGCCGCTGCCGATGGCGTCAAAT + ))%255CCF>>>>>>CCCCCCC65`IIII% ``` ] <i class="fa fa-frown-o" style="position:absolute;font-size:3em;left:8em;"></i> .blue[ ``` @PAIR-3 forward TCGCACTCAACGCCCTGCATATGACAAGAC + A64;##=#B9=AAAAAAAAAA9#:AB95%^ ``` ] <i class="fa fa-frown-o" style="position:absolute;font-size:3em;left:8em;"></i> .green[ ``` @PAIR-4 forward AAACTTCGTAGGTCCATTTGACAGCGTGCA + 6664%!!III^(=%3333^^d^d:#32333 ``` ] **`mysample_R1.fastq`** ] .pull-right[ .red[ ``` @PAIR-1 reverse AAGTTACCCTTAACAACTTAAGGGTTTTCA + fffddf`feedB`IABa)^%YBBBRTT\^d ``` ] .orange[ ``` @PAIR-2 reverse AGCAGAAGTCGATGATAATACGCGTCGTTT + IIIIIII^^IIId`?III%IIIGII>IIII ``` ] .blue[ ``` @PAIR-3 reverse AATCCATTTGTTCAACTCACAGTTTACCGT + 9C;=;=<9@4868>9:67AA<9>65<=>59 ``` ] **`mysample_R2.fastq`** ] ??? By cutting the yellow read only from the forward reads file, but leaving the other side of pair in the other file, an incorrect pairing is now assumed by downstream tools --- ## Choosing an Aligner - Each tool makes **different choices** during alignment - Choice of aligner may **affect downstream results** - Default options may not be best for your data! - Best tool for your data **depends on many factors** - Type of experiment (e.g. DNA, RNA, Bisulphite) - Sequencing platform - Compute resources vs sensitivity - Read characteristics (paired/single end, read length) .center[ .image-40[] ] .footnote[**Figure:** mapping of RNA-seq reads is different than DNA-seq] ??? Choice of mapper depends on your experiment - Some mappers are good for DNA but not RNA - Some mappers do well in highly rearranged genomes (e.g. cancer), others less so - Some mappers do well on some platforms but worse on others - e.g. Oxford Nanopore with its long reads but high error rates Or other factors - STAR needs a LOT of RAM - Do you need results fast? or accurate? (e.g. medical setting) FAQ: "Why not map RNA reads to the transcriptome?" - you can, and it is done, but then cannot find novel genes or alternative splicing FAQ: "Why not BLAST or BLAT?" - optimized for longer sequences - not base quality aware - too slow --- # Know your data! *“... there is no tool that outperforms all of the others in all the tests. Therefore, the end user should clearly specify [their] needs in order to choose the tool that provides the best results.”* - Hatem et al BMC Bioinformatics 2013, 14:184 .footnote[ [DOI: 10.1186/1471-2105-14-184](https://doi.org/10.1186/1471-2105-14-184) ] ??? Know the data you are working with and pick the right mapper and parameters for the job! Not an easy task.. --- class: top ## Mapping tools  .footnote[60+ different mappers, many comparison papers. Figure from [10.1093/bioinformatics/bts605](https://doi.org/10.1093/bioinformatics/bts605) ] ??? Many different tools available Different strengths and weaknesses, comparison table in link --- class: top # Mapping tools **Mapping tool** | **Uses** | **Characteristics** --- | --- | --- HISAT2 | DNA/RNA | Short reads. Based on [GCSA](https://doi.org/10.1109/TCBB.2013.2297101). [Reference](https://www.nature.com/articles/s41587-019-0201-4). RNASTAR | RNA | Short reads. Extremely fast. High sensitive and accuracy. Based on Maximal Mappable Prefixes (MMPs). [Reference](https://pubmed.ncbi.nlm.nih.gov/23104886/). BWA-MEM2 | DNA | Short reads. Twice as faster as BWA-MEM. Memory efficient. Based on [Burrows-Wheeler](https://academic.oup.com/bioinformatics/article/25/14/1754/225615). [Reference](https://arxiv.org/abs/1907.12931). Minimap2 | DNA/RNA | Long reads (PacBio and ONT). Extremely fast. Based on [DALIGN](https://link.springer.com/chapter/10.1007/978-3-662-44753-6_5) and [MHAP](https://www.nature.com/articles/nbt.3238). [Reference](https://doi.org/10.1093/bioinformatics/bty191). Bismark | DNA/RNA | Short reads. Bisulfite treated sequencing. Based on [GCSA](https://doi.org/10.1109/TCBB.2013.2297101). [Reference](https://pubmed.ncbi.nlm.nih.gov/21493656/). BBMap | DNA/RNA | Short and long reads (PacBio and ONT). Memory demanding. [Reference](https://bib.irb.hr/datoteka/773708.Josip_Maric_diplomski.pdf). Whisper 2 | DNA | Short reads. Indel sensitive. Variant-calling oriented. [Reference](https://academic.oup.com/bioinformatics/article/35/12/2043/5165374). S-conLSH | DNA | Long reads (ONT). High sensitivity and accuracy. [Reference](https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-020-03918-3). --- # File Formats --- # SAM/BAM file format  **SAM:** **S**equence **A**lignment **M**ap **BAM:** Binary (compressed) SAM; not human-readable --- # SAM/BAM file format  - Original read information (from FASTQ) plus mapping information - Position on reference, alignment, quality score, uniqueness, .. ??? Alignment given in CIGAR string. - in screenshot "37M" means 37 matches - in screenshot "18M2D19M" means 18 matches, then 2 deletions, then 19 matches --- class: top # Genome Browsers - Visualise aligned reads (BAM files)  .footnote[This is [IGV (Integrative Genome Browser)](https://software.broadinstitute.org/software/igv/) DOI: [10.1038/nbt.1754](https://doi.org/10.1038/nbt.1754)] ??? - Can zoom in and out, drag left and right, explore your sample - Zoom in for more information, mismatches, read information - Many different genome browsers exist --- class: top # Genome Browsers in Galaxy - [JBrowse](https://jbrowse.org/) <i class="fas fa-wrench" aria-hidden="true"></i><span class="visually-hidden">tool</span> Genome Browser as Galaxy tool .image-90[] .footnote[[JBrowse.org](https://jbrowse.org) DOI: [10.1186/s13059-016-0924-1](https://doi.org/10.1186/s13059-016-0924-1)] ??? Jbrowse tool builds up a small website for you, and pre-processes the reference genome into a more efficient format. If you wanted to share this with your colleagues, you could download this dataset and directly place it on your webserver. --- class: top # Genome Browsers in Galaxy - **External Genome Browsers** in Galaxy - BAM datasets in Galaxy have display links - [UCSC Genome Browser](https://genome-euro.ucsc.edu/cgi-bin/hgTracks), [Ensemble](https://www.ensembl.org), [IGV](https://software.broadinstitute.org/software/igv/), [IGB](https://bioviz.org/), [BAM.iobo](https://bam.iobio.io/home) 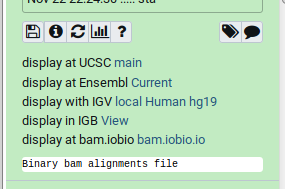 - Two different links for **IGV** - **local:** - Start IGV on your machine first - Then click link to automatically load data from Galaxy - **[Reference genome name]** (*"Human hg19"* in screenshot) - Downloads and starts IGV for you - Requires [Java web start](https://java.com/en/download/faq/java_webstart.xml) be installed on your machine ??? In the mapping hands-on tutorial you will use JBrowse and IGV --- ### <i class="fas fa-key" aria-hidden="true"></i><span class="visually-hidden">keypoints</span> Key points - Mapping is not trivial - There are many mapping tools, best choice depends on your data - Choice of mapper can affect downstream results - Know your data! - Genome browsers can be used to view aligned reads --- ## Thank You! This material is the result of a collaborative work. Thanks to the [Galaxy Training Network](https://training.galaxyproject.org) and all the contributors! <div markdown="0"> <div class="contributors-line"> <table class="contributions"> <tr> <td><abbr title="These people wrote the bulk of the tutorial, they may have done the analysis, built the workflow, and wrote the text themselves.">Author(s)</abbr></td> <td> <a href="/training-material/hall-of-fame/joachimwolff/" class="contributor-badge contributor-joachimwolff"><img src="https://avatars.githubusercontent.com/joachimwolff?s=36" alt="Joachim Wolff avatar" width="36" class="avatar" /> Joachim Wolff</a><a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-shiltemann"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /> Saskia Hiltemann</a><a href="/training-material/hall-of-fame/EngyNasr/" class="contributor-badge contributor-EngyNasr"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/EngyNasr?s=36" alt="Engy Nasr avatar" width="36" class="avatar" /> Engy Nasr</a><a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-gallardoalba"><img src="/training-material/assets/images/orcid.png" alt="orcid logo" width="36" height="36"/><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /> Cristóbal Gallardo</a> </td> </tr> <tr class="reviewers"> <td><abbr title="These people reviewed this material for accuracy and correctness">Reviewers</abbr></td> <td> <a href="/training-material/hall-of-fame/hexylena/" class="contributor-badge contributor-badge-small contributor-hexylena"><img src="https://avatars.githubusercontent.com/hexylena?s=36" alt="Helena Rasche avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/gallardoalba/" class="contributor-badge contributor-badge-small contributor-gallardoalba"><img src="https://avatars.githubusercontent.com/gallardoalba?s=36" alt="Cristóbal Gallardo avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/abretaud/" class="contributor-badge contributor-badge-small contributor-abretaud"><img src="https://avatars.githubusercontent.com/abretaud?s=36" alt="Anthony Bretaudeau avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/bgruening/" class="contributor-badge contributor-badge-small contributor-bgruening"><img src="https://avatars.githubusercontent.com/bgruening?s=36" alt="Björn Grüning avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/shiltemann/" class="contributor-badge contributor-badge-small contributor-shiltemann"><img src="https://avatars.githubusercontent.com/shiltemann?s=36" alt="Saskia Hiltemann avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/nsoranzo/" class="contributor-badge contributor-badge-small contributor-nsoranzo"><img src="https://avatars.githubusercontent.com/nsoranzo?s=36" alt="Nicola Soranzo avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/bebatut/" class="contributor-badge contributor-badge-small contributor-bebatut"><img src="https://avatars.githubusercontent.com/bebatut?s=36" alt="Bérénice Batut avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/anuprulez/" class="contributor-badge contributor-badge-small contributor-anuprulez"><img src="https://avatars.githubusercontent.com/anuprulez?s=36" alt="Anup Kumar avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/willdurand/" class="contributor-badge contributor-badge-small contributor-willdurand"><img src="https://avatars.githubusercontent.com/willdurand?s=36" alt="William Durand avatar" width="36" class="avatar" /></a><a href="/training-material/hall-of-fame/njall/" class="contributor-badge contributor-badge-small contributor-njall"><img src="https://avatars.githubusercontent.com/njall?s=36" alt="Niall Beard avatar" width="36" class="avatar" /></a></td> </tr> </table> </div> </div> <div style="display: flex;flex-direction: row;align-items: center;justify-content: center;"> <img src="/training-material/assets/images/GTNLogo1000.png" alt="Galaxy Training Network" style="height: 100px;"/> <div> <div> <img class="funder-avatar" src="https://avatars.githubusercontent.com/gallantries" alt="Logo"> </div> <div> </div> </div> <div> <div> <img class="funder-avatar" src="https://avatars.githubusercontent.com/elixir-europe" alt="Logo"> </div> <div> </div> </div> </div> Tutorial Content is licensed under <a rel="license" href="http://creativecommons.org/licenses/by/4.0/">Creative Commons Attribution 4.0 International License</a>.<br/>