Genome Assembly of a bacterial genome (MRSA) sequenced using Illumina MiSeq Data
| Author(s) |
|
| Editor(s) |
|
| Reviewers |
|
OverviewQuestions:
Objectives:
How to check the quality of the MiSeq data?
How to perform an assembly of a bacterial genome with MiSeq data?
How to check the quality of an assembly?
Requirements:
Run tools to evaluate sequencing data on quality and quantity
Process the output of quality control tools
Improve the quality of sequencing data
Run a tool to assemble a bacterial genome using short reads
Run tools to assess the quality of an assembly
Understand the outputs of tools to assess the quality of an assembly
Time estimation: 2 hoursSupporting Materials:Published: Mar 24, 2021Last modification: Jan 5, 2026License: Tutorial Content is licensed under Creative Commons Attribution 4.0 International License. The GTN Framework is licensed under MITpurl PURL: https://gxy.io/GTN:T00036rating Rating: 4.5 (4 recent ratings, 20 all time)version Revision: 18
Sequencing (determining of DNA/RNA nucleotide sequence) is used all over the world for all kinds of analysis. The product of these sequencers are reads, which are sequences of detected nucleotides. Depending on the technique these have specific lengths (30-500bp) or using Oxford Nanopore Technologies sequencing have much longer variable lengths.
Comment: Illumina MiSeq sequencingIllumina MiSeq sequencing is based on sequencing by synthesis. As the name suggests, fluorescent labels are measured for every base that bind at a specific moment at a specific place on a flow cell. These flow cells are covered with oligos (small single strand DNA strands). In the library preparation the DNA strands are cut into small DNA fragments (differs per kit/device) and specific pieces of DNA (adapters) are added, which are complementary to the oligos. Using bridge amplification large amounts of clusters of these DNA fragments are made. The reverse string is washed away, making the clusters single stranded. Fluorescent bases are added one by one, which emit a specific light for different bases when added. This is happening for whole clusters, so this light can be detected and this data is basecalled (translation from light to a nucleotide) to a nucleotide sequence (Read). For every base a quality score is determined and also saved per read. This process is repeated for the reverse strand on the same place on the flow cell, so the forward and reverse reads are from the same DNA strand. The forward and reversed reads are linked together and should always be processed together!
For more information watch this video from Illumina
In this training we will build an assembly of a bacterial genome, from data produced in “Complete Genome Sequences of Eight Methicillin-Resistant Staphylococcus aureus Strains Isolated from Patients in Japan” Hikichi et al. 2019:
Methicillin-resistant Staphylococcus aureus (MRSA) is a major pathogen causing nosocomial infections, and the clinical manifestations of MRSA range from asymptomatic colonization of the nasal mucosa to soft tissue infection to fulminant invasive disease. Here, we report the complete genome sequences of eight MRSA strains isolated from patients in Japan.
AgendaIn this tutorial, we will cover:
Galaxy and data preparation
Any analysis should get its own Galaxy history. So let’s start by creating a new one and get the data into it.
Hands On: History creation
Create a new history for this analysis
To create a new history simply click the new-history icon at the top of the history panel:
Rename the history
- Click on galaxy-pencil (Edit) next to the history name (which by default is “Unnamed history”)
- Type the new name
- Click on Save
- To cancel renaming, click the galaxy-undo “Cancel” button
If you do not have the galaxy-pencil (Edit) next to the history name (which can be the case if you are using an older version of Galaxy) do the following:
- Click on Unnamed history (or the current name of the history) (Click to rename history) at the top of your history panel
- Type the new name
- Press Enter
Now, we need to import the data: 2 FASTQ files containing the reads from the sequencer.
Hands On: Import datasets
Import the files from Zenodo or from Galaxy shared data libraries:
https://zenodo.org/record/10669812/files/DRR187559_1.fastqsanger.bz2 https://zenodo.org/record/10669812/files/DRR187559_2.fastqsanger.bz2
- Copy the link location
Click galaxy-upload Upload at the top of the activity panel
- Select galaxy-wf-edit Paste/Fetch Data
Paste the link(s) into the text field
Press Start
- Close the window
As an alternative to uploading the data from a URL or your computer, the files may also have been made available from a shared data library:
- Go into Libraries (left panel)
- Navigate to the correct folder as indicated by your instructor.
- On most Galaxies tutorial data will be provided in a folder named GTN - Material –> Topic Name -> Tutorial Name.
- Select the desired files
- Click on Add to History galaxy-dropdown near the top and select as Datasets from the dropdown menu
In the pop-up window, choose
- “Select history”: the history you want to import the data to (or create a new one)
- Click on Import
Rename the datasets to remove
.fastqsanger.bz2and keep only the sequence run ID (DRR187559_1andDRR187559_2)
- Click on the galaxy-pencil pencil icon for the dataset to edit its attributes
- In the central panel, change the Name field to
DRR187559_1- Click the Save button
Create a paired collection named
Paired Reads
- Click on galaxy-selector Select Items at the top of the history panel
- Check all the datasets in your history you would like to include
Click n of N selected and choose Advanced Build List
You are in the collection building wizard. Choose List of Paired Datasets and click ‘Next’ button at the right bottom corner.
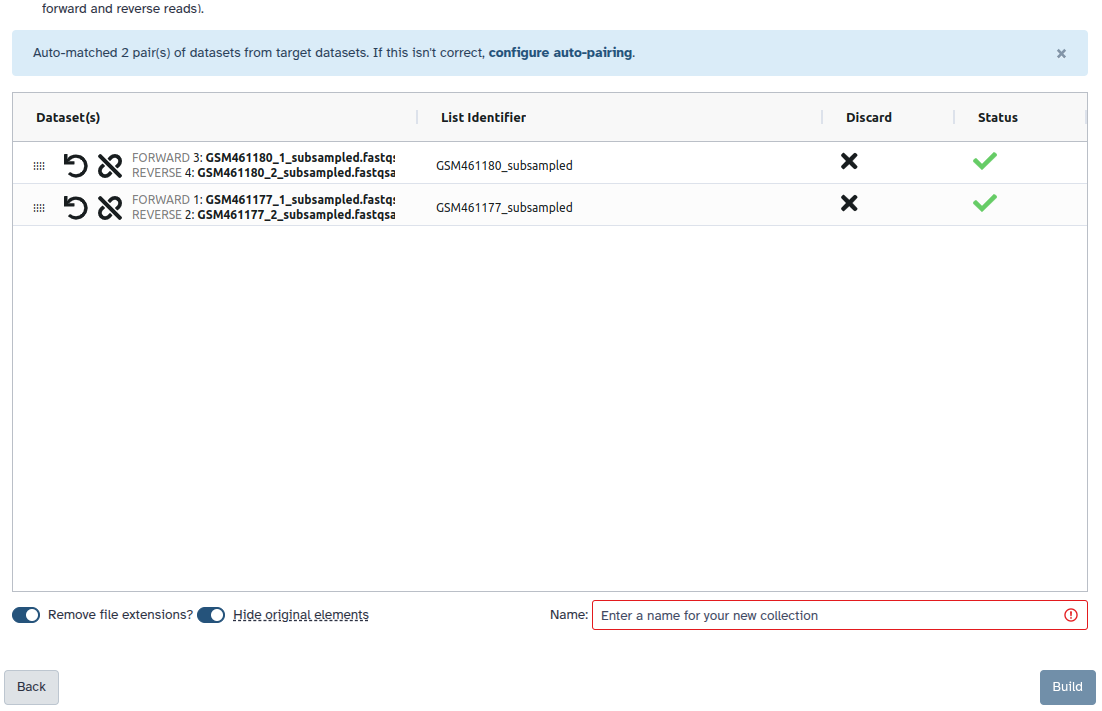
Check and configure auto-pairing. Commonly matepairs have suffix
_1and_2or_R1and_R2. Click on ‘Next’ at the bottom.
- Edit the List Identifier as required.
- Enter a name for your collection
- Click Build to build your collection
- Click on the checkmark icon at the top of your history again
Tag the collection
#unfiltered
- Click on the collection in your history to view it
- Click on Edit galaxy-pencil next to the collection name at the top of the history panel
- Click on Add Tags galaxy-tags
- Add a tag starting with
#
- Tags starting with
#will be automatically propagated to the outputs any tools using this dataset.- Click Save galaxy-save
- Check that the tag appears below the collection name
View galaxy-eye the renamed files in the collection
The datasets are both FASTQ files.
Question
- What are the 4 main features of each read in a FASTQ file.
- What does the
_1and_2mean in your filenames?
The following:
- A
@followed by a name and sometimes information of the read- A nucleotide sequence
- A
+(optional followed by the name)- The quality score per base of nucleotide sequence (Each symbol represents a quality score, which will be explained later)
Forward and reverse reads, by convention, are labelled
_1and_2, but they might also be_f/_ror_r1/_r2.
Quality Control
During sequencing, errors are introduced, such as incorrect nucleotides being called. These are due to the technical limitations of each sequencing platform. Sequencing errors might bias the analysis and can lead to a misinterpretation of the data. Adapters may also be present if the reads are longer than the fragments sequenced and trimming these may improve the number of reads mapped. Sequence quality control is therefore an essential first step in any analysis.
Assessing the quality by hand would be too much work. That’s why tools like NanoPlot or Falco are made, as they generate a summary and plots of the data statistics. NanoPlot is mainly used for long-read data, like ONT and PACBIO and Falco for short read, like Illumina and Sanger. Falco is an efficiency-optimized rewrite of FastQC. You can read more in our dedicated Quality Control Tutorial.
Before doing any assembly, the first questions we should ask about the input reads include:
- What is the coverage of my genome?
- How good are my reads?
- Do I need to ask/perform for a new sequencing run?
- Is it suitable for the analysis I need to do?
Hands On: Quality Control
- Falco ( Galaxy version 1.2.4+galaxy0) with the following parameters:
- param-collection “Raw read data from your current history”:
Paired Reads- Inspect the webpage outputs
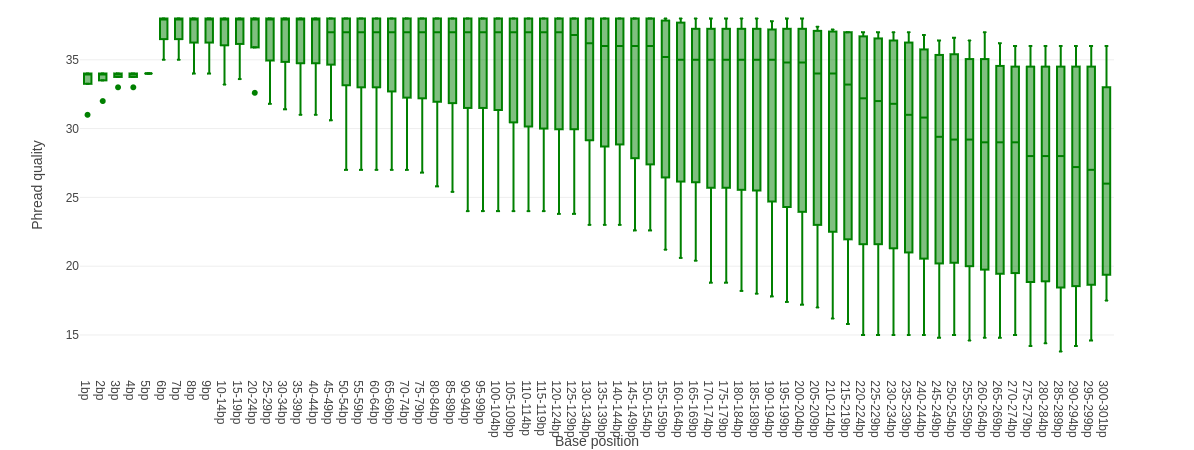
Falco combines quality statistics from all separate reads and combines them in plots. An important plot is the Per base sequence quality.
| DRR187559_1 | DRR187559_2 |
|---|---|
 |
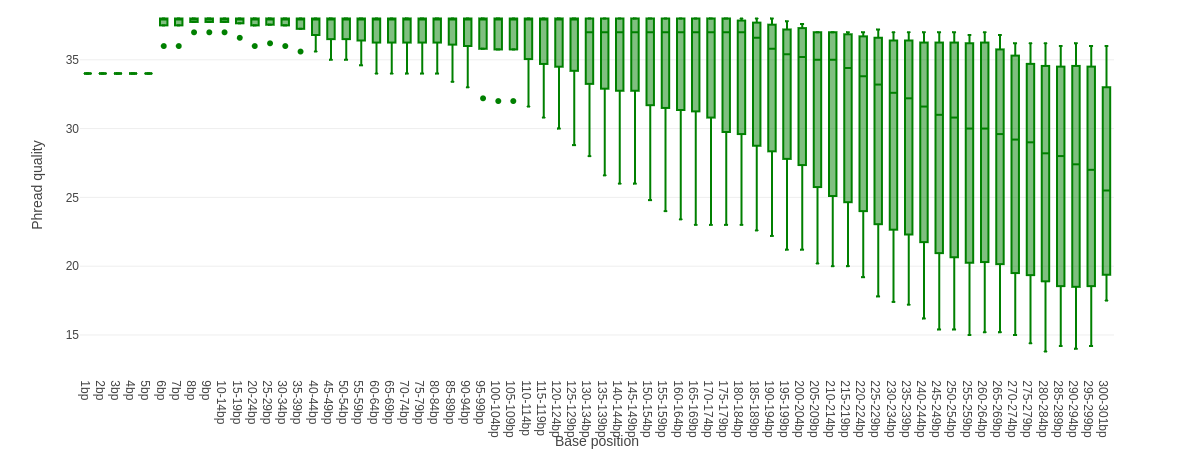 |
Here you have the reads sequence length on the x-axes against the quality score (Phred-score) on the y-axis. The colour of the beams indicates three quality levels:
- Green = good quality,
- Yellow = mediocre quality, and
- Red = bad quality.
For each position, a boxplot is drawn with:
- the median value, represented by the central intense coloured line
- the inter-quartile range (25-75%), represented by the geen, yellow or red box
- the 10% and 90% values in the upper and lower whiskers
For Illumina data it is normal that the first few bases are of some lower quality and how longer the reads get the worse the quality becomes. This is often due to signal decay or phasing during the sequencing run.
Before doing any assembly, the first questions we should ask about the input reads include:
- What is the coverage of my genome?
- How good are my reads?
- Do I need to ask/perform for a new sequencing run?
- Is it suitable for the analysis I need to do?
Depending on the analysis it could be possible that a certain quality or length is needed. In this case we are going to trim the data using fastp (Chen et al. 2018):
-
Trim the start and end of the reads if those fall below a quality score of 20
Different trimming tools have different algorithms for deciding when to cut but trimmomatic will cut based on the quality score of one base alone. Trimmomatic starts from each end, and as long as the base is below 20, it will be cut until it reaches one greater than 20. A sliding window trimming will be performed where if the average quality of 4 bases drops below 20, the read will be truncated there.
-
Filter for reads to keep only reads with at least 30 bases: Anything shorter will complicate the assembly
Hands On: Quality improvement
- fastp ( Galaxy version 1.0.1+galaxy3) with the following parameters:
- “Single-end or paired reads”:
Paired Collection
- “Select paired collection(s)”:
Paired Reads- In “Filter Options”:
- In “Length filtering Options”:
- Length required:
30- In “Read Modification Options”:
- In “Per read cuitting by quality options”:
- Cut by quality in front (5’):
Yes- Cut by quality in tail (3’):
Yes- Cutting window size:
4- Cutting mean quality:
20- In “Output Options”:
- “Output JSON report”:
Yes- Edit the tags of the fastp FASTQ outputs to
- Remove the
#unfilteredtag- Add a new tag
#filtered
fastp generates also a report, similar to Falco, useful to compare the impact of the trimming and filtering.
QuestionLooking at fastp HTML report
- How did the average read length change before and after filtering?
- Did trimming improve the mean quality scores?
- Did trimming affect the GC content?
- Is this data ok to assemble? Do we need to re-sequence it?
- Read lengths went down more significantly:
- Before filtering: 190bp, 221bp
- After filtering: 189bp, 219bp
- It increased the percentage of Q20 and Q30 bases (bases with quality score above 20 and 30 respectively)
- No, it did not. If it had, that would be unexpected.
- This data looks OK. The number of short reads in R1 is not optimal but assembly should partially work but not the entire, closed genome.
Assembly
Now that the quality of the reads is determined and the data is filtered and/or trimmed, we can try to assemble the reads together to build longer sequences.
There are many tools that create assembly for short-read data, e.g. SPAdes (Prjibelski et al. 2020), Abyss (Simpson et al. 2009). In this tutorial, we use Shovill. Shovill is a SPAdes-based genome assembler, improved to work faster and only for smaller (bacterial) genomes.
Assembly
Comment: Results may varyYour results may be slightly different from the ones presented in this tutorial due to differing versions of tools, reference data, external databases, or because of stochastic processes in the algorithms.
Hands On: Assembly using Shovill
- Shovill ( Galaxy version 1.1.0+galaxy1) with the following parameters:
- “Input reads type, collection or single library”:
Paired Collection
- “Paired collection”: fastp
Paired-end output- In “Advanced options”:
- “Estimated genome size”:
2914567
*Shovill generates 3 outputs:
- A log file
-
A FASTA file with contigs, i.e. contiguous length of genomic sequences in which bases are known to a high degree of certainty.
QuestionInspect the
Contigsfile- How big is the first contig?
- What is the coverage of your biggest (first) contig?
Both of these can be found in the header line of the Contigs produced by Shovill
The results can differ from this example, because Shovill can differ a bit per assembly
- 589,438 bp, almost 6 times lower than the estimated genome size
- 17.7
-
A file with the contig graph or assembly graph
The assembly graph is more information-rich because it not only contains the sequences of all assembled fragments (including the ones shorter than the threshold length defined for inclusion of the fragments into the FASTA output), but also indicates the relative average coverage of the fragments by sequenced reads and how some of the fragments could potentially be bridged after resolving ambiguities manually.
Assembly inspection
Th assembly graph format takes some getting used to before you can make sense out of the information it provides. This issue can be alleviated through the use of Bandage (Wick et al. 2015), a package for exploring assembly graphs through summary reports and visualizations of their contents.
Hands On: Assembly inspection
- Bandage Info ( Galaxy version 2022.09+galaxy2) with the following parameters:
- param-file “Graphical Fragment Assembly”: Shovill
Contig Graph- “Produce jpg, png or svg file?”:
.jpg
Let’s look at the assembly statistics
Question
- What is the number of dead end?
- What is the number of nodes?
- Only 2
- 131: a bit disapointing as it should be encoded in one contig
Let’s now visualize the graph.
Hands On: Assembly inspection
- Bandage Image ( Galaxy version 2022.09+galaxy4) with the following parameters:
- param-file “Graphical Fragment Assembly”: Shovill
Contig Graph

QuestionFirst read this page in the Bandage wiki to help understand what the graph means.
What do you think of this assembly? Is it useful? Is it good enough?
This is a very messy assembly, with a lot of potential paths through the sequence. We cannot feel confident in the output FASTA file (as it is much smaller than the expected 2.9Mbp). In real life we might consider doing a hybrid assembly with Nanopore or other long read data to help resolve these issues.
Assembly Evaluation
To evaluate the assembly, we use also Quast (Gurevich et al. 2013) (QUality ASsessment Tool), a tool providing quality metrics for assemblies. This tool can be used to compare multiple assemblies, can take an optional reference file as input to provide complementary metrics, etc
Hands On: Quality Control of assembly using Quast
- Quast ( Galaxy version 5.2.0+galaxy1) with the following parameters:
- Assembly mode?:
Co-assembly
- “Use customized names for the input files?”:
No, use dataset names
- param-file “Contigs/scaffolds file”: Shovill
Contigsoutput
QUAST outputs assembly metrics as an HTML file with metrics and graphs.
This fasta file contains
Question
- How many contigs is there?
- What is the total length of all contigs?
- What is you GC content?
- 30 contigs, meaning the chromosome is separated over multiple contigs. These contigs can also contain (parts of) plasmids.
- 2,911,349 (Total length (>= 0 bp)). Not far from the estimated genome size: 2,914,567
- The GC content for our assembly was 32.77%. For comparison MRSA Isolate HC1335 Strain GC% is 32.89%.
The total length and the GC content of the assembly are coherent with expectations.
Conclusion
In this tutorial, we prepared short reads, assembled them, and inspect the produced assembly for its quality. The assembly, even if uncomplete, is reasonable good to be used in downstream analysis, like AMR gene detection