Maria Doyle
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Topic: Transcriptomics
Tutorials
- Assembly / An Introduction to Genome Assembly 🧐
- Assembly / De Bruijn Graph Assembly 🧐
- Assembly / Making sense of a newly assembled genome 🧐
- Genome Annotation / CRISPR screen analysis ✍️
- Genome Annotation / Genome annotation with Prokka 🧐
- Using Galaxy and Managing your Data / Extracting Workflows from Histories 🧐
- Using Galaxy and Managing your Data / Using dataset collections 🧐
- Variant Analysis / Microbial Variant Calling 🧐
- Variant Analysis / Calling very rare variants 🧐
- Transcriptomics / 3: RNA-seq genes to pathways ✍️
- Transcriptomics / Visualization of RNA-Seq results with heatmap2 ✍️
- Transcriptomics / Reference-based RNA-Seq data analysis ✍️ 🧐
- Transcriptomics / 1: RNA-Seq reads to counts ✍️ 🧐
- Transcriptomics / Visualization of RNA-Seq results with CummeRbund 🧐
- Transcriptomics / Network analysis with Heinz 🧐
- Transcriptomics / 2: RNA-seq counts to genes ✍️ 🧐
- Transcriptomics / GO Enrichment Analysis ✍️ 🧐
- Transcriptomics / Visualization of RNA-Seq results with Volcano Plot in R ✍️
- Transcriptomics / Visualization of RNA-Seq results with Volcano Plot ✍️
- Epigenetics / ATAC-Seq data analysis ✍️
- Epigenetics / Identification of the binding sites of the Estrogen receptor 🧐
- Epigenetics / Identification of the binding sites of the T-cell acute lymphocytic leukemia protein 1 (TAL1) 🧐
- Introduction to Galaxy Analyses / A short introduction to Galaxy 🧐
- Introduction to Galaxy Analyses / From peaks to genes 🧐
- Introduction to Galaxy Analyses / Introduction to Genomics and Galaxy 🧐
- Introduction to Galaxy Analyses / Galaxy Basics for genomics 🧐
- Sequence analysis / Mapping 🧐
- Sequence analysis / Quality Control ✍️ 🧐
Slides
- Galaxy Server administration / Advanced customisation of a Galaxy instance 🧐
- Genome Annotation / Introduction to CRISPR screen analysis ✍️
- Using Galaxy and Managing your Data / Getting data into Galaxy ✍️
FAQs
Video Recordings
- Genome Annotation / Introduction to CRISPR screen analysis 💬
- Genome Annotation / CRISPR screen analysis 💬 🗣
- Transcriptomics / Visualization of RNA-Seq results with Volcano Plot in R 💬 🗣
- Transcriptomics / Visualization of RNA-Seq results with Volcano Plot 💬 🗣
Events
- Bioconductor Carpentries workshop: Analysis and Interpretation of Bulk RNA-Seq Data 🎪
- Bioconductor North American conference (BioC2024) 🎪
- EuroBioC2024 Carpentries workshops 🎪
GitHub Activity
github Issues Reported
49 Merged Pull Requests
See all of the github Pull Requests and github Commits by Maria Doyle.
-
 Fix zenodo links
genome-annotation
Fix zenodo links
genome-annotation -
 RNA-seq reads counts: small update
RNA-seq reads counts: small update
-
 Crispr slides fix
Crispr slides fix
-
 CRISPR: Add speaker notes to slide
CRISPR: Add speaker notes to slide
-
 CRISPR finalise
CRISPR finalise
Reviewed 13 PRs
We love our community reviewing each other's work!
-
 Update Transcriptomics Reference-based tutorial
transcriptomicstemplate-and-tools
Update Transcriptomics Reference-based tutorial
transcriptomicstemplate-and-tools -
 Quality control: update tutorial to include FASTQE
CoFest
Quality control: update tutorial to include FASTQE
CoFest -
 Reconfiguration of some content in mouse RNA-seq tutorials
Reconfiguration of some content in mouse RNA-seq tutorials
-
 Ref-based RNA-Seq: some fixes, reorganization, QC checks on mapped reads
transcriptomics
Ref-based RNA-Seq: some fixes, reorganization, QC checks on mapped reads
transcriptomics -
 RNA-seq limma-voom rework
transcriptomicsnew tutorial
RNA-seq limma-voom rework
transcriptomicsnew tutorial
News
Next GTN CoFest May 20, 2021
18 March 2021
Every 3 months we organise one day dedicated to the GTN community! Thursday, May 20th is the next GTN CoFest. We will be working on the training materials, have discussions with the global Galaxy traning community. Do you teach with Galaxy? Want to learn how to add your tutorial to the GTN? New to the community and just want to learn more? Everybody is welcome!
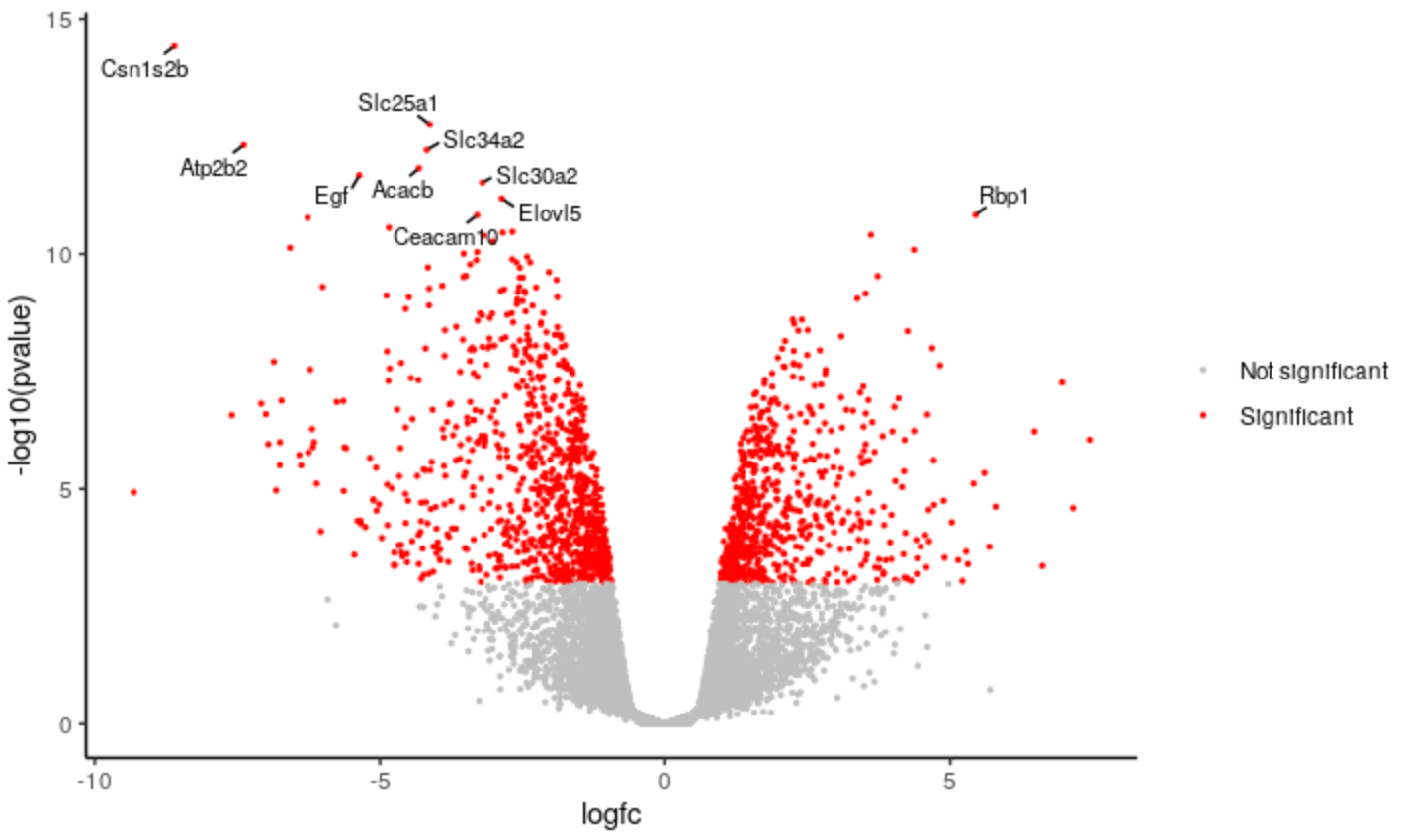
New Tutorial: Visualization of RNA-Seq results with Volcano Plot in R
26 June 2021
The Volcano plot tutorial introduced volcano plots and showed how they can be easily generated with the Galaxy Volcano plot tool. This new tutorial shows how you can customise a plot using the R script output from the tool and RStudio in Galaxy. A short video for the tutorial is also available on YouTube, created for the GCC2021 Training week.
Happy plotting!