Mehmet Tekman
Affiliations
Former Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Learning Pathway: Bioinformatics Projects: Using deconvolution to get new insights from old bulk RNA-seq data
Tutorials
- Transcriptomics / 1: RNA-Seq reads to counts 🧐
- Transcriptomics / Reference-based RNA-Seq data analysis 🧐
- Using Galaxy and Managing your Data / SRA Aligned Read Format to Speed Up SARS-CoV-2 data Analysis 🧐
- Introduction to Galaxy Analyses / How to reproduce published Galaxy analyses 🧐
- Introduction to Galaxy Analyses / A short introduction to Galaxy 🧐
- Introduction to Galaxy Analyses / NGS data logistics 🧐
- Teaching and Hosting Galaxy training / Training Infrastructure as a Service 🧐
- Sequence analysis / Mapping 🧐
- Contributing to the Galaxy Training Material / Creating content in Markdown 🧐
- Epigenetics / Hi-C analysis of Drosophila melanogaster cells using HiCExplorer 🧐
- Single Cell / Analysis of plant scRNA-Seq Data with Scanpy ✍️ 🧐
- Single Cell / Understanding Barcodes ✍️ 🧐
- Single Cell / Filter, plot and explore single-cell RNA-seq data with Scanpy (Python) 🧐
- Single Cell / Importing files from public atlases 🧐
- Single Cell / Pre-processing of 10X Single-Cell ATAC-seq Datasets 🧐
- Single Cell / Downstream Single-cell RNA analysis with RaceID ✍️ 🧐
- Single Cell / Matrix Exchange Format to ESet | Creating a single-cell RNA-seq reference dataset for deconvolution ✍️ 🧐
- Single Cell / Filter, plot and explore single-cell RNA-seq data with Scanpy 🧐
- Single Cell / Pre-processing of Single-Cell RNA Data ✍️ 🧐
- Single Cell / Inferring single cell trajectories with Scanpy ✍️ 🧐
- Single Cell / Bulk matrix to ESet | Creating the bulk RNA-seq dataset for deconvolution ✍️ 🧐
- Single Cell / Converting between common single cell data formats ⚙️ 🧐
- Single Cell / Inferring single cell trajectories with Monocle3 🧐
- Single Cell / Single-cell quality control with scater 🧐
- Single Cell / Scanpy Parameter Iterator 🧐
- Single Cell / Removing the effects of the cell cycle 🧐
- Single Cell / Generating a single cell matrix using Alevin and combining datasets (bash + R) 📝 🧐
- Single Cell / Combining single cell datasets after pre-processing 🧐
- Single Cell / Bulk RNA Deconvolution with MuSiC ✍️ 🧐
- Single Cell / Inferring single cell trajectories with Monocle3 (R) 🧐
- Single Cell / Inferring single cell trajectories with Scanpy (Python) ✍️ 🧐
- Single Cell / Filter, plot, and explore single cell RNA-seq data with Seurat (R) 📝 🧐
- Single Cell / Pre-processing of 10X Single-Cell RNA Datasets ✍️ 🧐
- Single Cell / Converting NCBI Data to the AnnData Format 📝 🧐
- Single Cell / Comparing inferred cell compositions using MuSiC deconvolution ✍️ 🧐
- Single Cell / Generating a single cell matrix using Alevin 🧐
- Single Cell / Clustering 3K PBMCs with Scanpy ✍️ 🧐
- Ecology / Production d'indicateurs champs de bloc 🧐
Slides
- Introduction to Galaxy Analyses / A Short Introduction to Galaxy 🧐
- Teaching and Hosting Galaxy training / Workshop Kickoff 🧐
- Galaxy Server administration / Galaxy from an administrator's point of view 🧐
- Single Cell / Plates, Batches, and Barcodes ✍️ 🧐
- Single Cell / Dealing with Cross-Contamination in Fixed Barcode Protocols ✍️
- Single Cell / An introduction to scRNA-seq data analysis ✍️ 🧐
- Single Cell / Trajectory analysis 🧐
- Single Cell / Automated Cell Annotation 🧐
- Single Cell / Clustering 3K PBMCs with Scanpy ✍️ 🧐
- Introduction to Galaxy Analyses / Una breve introducción a Galaxy 🧐
- Single Cell / Introducción al análisis de datos de scRNA-seq ✍️ 🧐
- Single Cell / Una introducción al análisis de datos scRNA-seq ✍️ 🧐
FAQs
- Why is Alevin is not working?
- Why is Alevin is not working?
- Why is amplification more of an issue in scRNA-seq than RNA-seq?
- AnnData Import/ AnnData Manipulate not working?
- Importieren über Links ✍️
- Importing via links
- My Scanpy FindMarkers step is giving me an empty table
- On Scanpy PlotEmbed, the tool is failing
- On the Scanpy PlotEmbed step, my object doesn’t have Il2ra or Cd8b1 or Cd8a etc.
- How do I know what protocol my data was sequenced with?
- Are Barcodes always on R1 and Sequence data on R2?
- Can RNA-seq techniques be applied to scRNA-seq?
- The UMAP Plots errors out sometimes?
- Are UMIs not actually unique?
Video Recordings
- Single Cell / An introduction to scRNA-seq data analysis 💬
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads 💬
- Proteomics / EncyclopeDIA 💬
- Single Cell / Analysis of plant scRNA-Seq Data with Scanpy 💬 🗣
- Single Cell / Bulk RNA Deconvolution with MuSiC 💬
GitHub Activity
github Issues Reported
57 Merged Pull Requests
See all of the github Pull Requests and github Commits by Mehmet Tekman.
-
 PostmarketOS Galaxy Joke Post
news
PostmarketOS Galaxy Joke Post
news -
 Add News/Event: helper bots event
template-and-toolsnews
Add News/Event: helper bots event
template-and-toolsnews -
 Add UseGalaxy Help Forum to Matrix channel Bot
template-and-tools
Add UseGalaxy Help Forum to Matrix channel Bot
template-and-tools -
 MuSiC Tutorial
MuSiC Tutorial
-
 Fix to STARsolo parameters for use with 2.7.8a
Fix to STARsolo parameters for use with 2.7.8a
Reviewed 41 PRs
We love our community reviewing each other's work!
-
 SnapATAC2 tutorial: update data-library.yaml
single-cell
SnapATAC2 tutorial: update data-library.yaml
single-cell -
 Line edits by author for Filter Plot and Explore RStudio
single-cell
Line edits by author for Filter Plot and Explore RStudio
single-cell -
 PostmarketOS Galaxy Joke Post
news
PostmarketOS Galaxy Joke Post
news -
 Add variables for usegalaxy.star servers
adminintroductiontranscriptomicstemplate-and-toolsgalaxy-interface
Add variables for usegalaxy.star servers
adminintroductiontranscriptomicstemplate-and-toolsgalaxy-interface -
 misc e+/metadata
template-and-toolscontributing
misc e+/metadata
template-and-toolscontributing
News
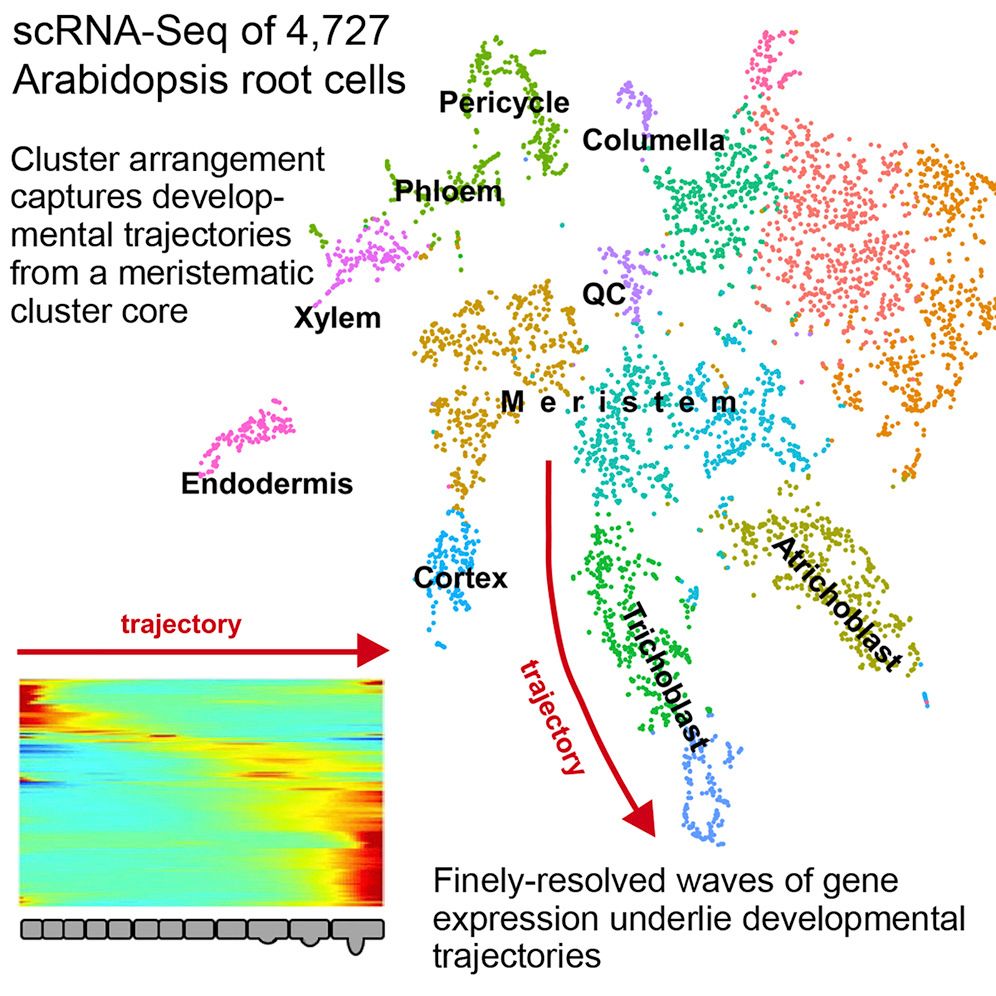
New Topic: Single Cell Analysis!
18 November 2022
Single-cell analysis now has it’s own topic! These tutorials were previously part of the transcriptomics topic, but due to the amazing efforts by
Wendi Bacon,
Mehmet Tekmanand others, we now have so many single-cell analysis tutorials that they deserve their own dedicated topic! From introductory slides and practicals, to case study tutorials generating cell clusters and trajectories from raw sequencing files, and even a growing subtopic of smaller tips and tricks for adapting analysis to user needs, the single cell topic either has what you need or is working on it. Come learn or get involved!
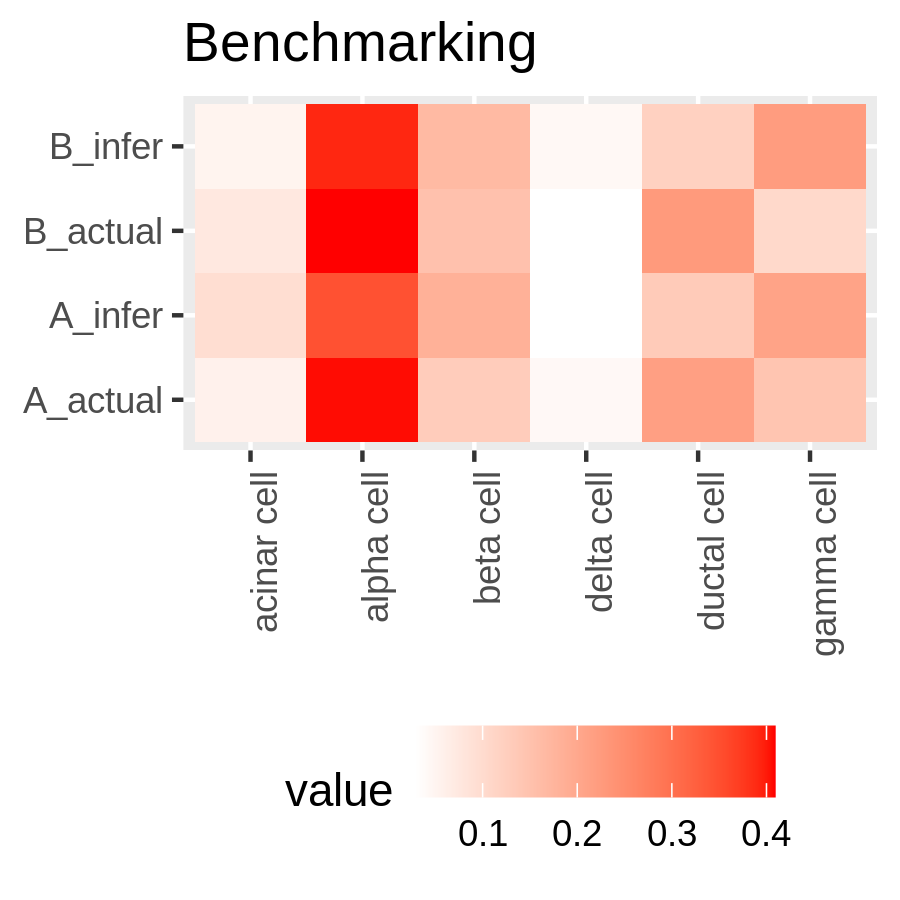
New Tutorial Suite: Deconvolution with MuSiC, from public data to disease interrogation!
29 November 2022
The still new and shiny single-cell analysis topic now boasts a deconvolution tutorial suite! What does deconvolution do you ask? Well, in this context, it infers cell proportions from bulk RNA-seq data. You heard that correctly - instead of expensive new single-cell experiments, you can re-analyse old bulk RNA-seq data and estimate cell proportions. All you need is a reasonably good single cell dataset to use as a reference and you’re good to go! The tutorial suite shows you how to build your reference from publicly available single cell data, and apply analysis to some publicly available bulk RNA-seq data.

Galaxy Single-cell Community: Year in Review
22 December 2023
🚀Embarking on a cosmic journey, the Galaxy Single-cell Community has clustered together to unveil a constellation of tools, making strides in RNA-stellar discoveries and creating out-of-this-world workflows. With a commitment to battling work duplication across the multiverse, this community is boldly charting a course for global domination, proving that when it comes to bioinformatics, the Galaxy is the limit!✨