Cristóbal Gallardo
Former Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Tutorials
- Variant Analysis / M. tuberculosis Variant Analysis 🧐
- Genome Annotation / From small to large-scale genome comparison 🧐
- Genome Annotation / Genome annotation with Funannotate 🧐
- Genome Annotation / Masking repeats with RepeatMasker 🧐
- Assembly / Large genome assembly and polishing 🧐
- Assembly / Using the VGP workflows to assemble a vertebrate genome with HiFi and Hi-C data ✍️ 🧐
- Assembly / Genome Assembly of MRSA from Oxford Nanopore MinION data (and optionally Illumina data) 🧐
- Assembly / Chloroplast genome assembly 🧐
- Assembly / Genome Assembly of a bacterial genome (MRSA) sequenced using Illumina MiSeq Data 🧐
- Assembly / Genome assembly using PacBio data 🧐
- Assembly / Genome Assembly Quality Control 🧐
- Assembly / An Introduction to Genome Assembly 🧐
- Assembly / Vertebrate genome assembly using HiFi, Bionano and Hi-C data - Step by Step ✍️ 🧐
- Assembly / ERGA post-assembly QC ✍️ 🧐
- Synthetic Biology / Generating theoretical possible pathways for the production of Lycopene in E.Coli using Retrosynthesis tools 🧐
- Transcriptomics / Network analysis with Heinz 🧐
- Transcriptomics / Reference-based RNA-Seq data analysis 🧐
- Transcriptomics / Genome-wide alternative splicing analysis ✍️ 🧐
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana ✍️ 🧐
- Using Galaxy and Managing your Data / Group tags for complex experimental designs 🧐
- Using Galaxy and Managing your Data / Rule Based Uploader 🧐
- Using Galaxy and Managing your Data / Understanding Galaxy history system 📝
- Introduction to Galaxy Analyses / Galaxy Basics for genomics 🧐
- Introduction to Galaxy Analyses / A short introduction to Galaxy 🧐
- Introduction to Galaxy Analyses / Introduction to Genomics and Galaxy ✍️ 🧐
- Development in Galaxy / Contributing to BioBlend as a developer 🧐
- Imaging / Object tracking using CellProfiler 🧐
- Metabolomics / Mass spectrometry: GC-MS analysis with the metaMS package 🧐
- Microbiome / Taxonomic Profiling and Visualization of Metagenomic Data 🧐
- Microbiome / Identification of the micro-organisms in a beer using Nanopore sequencing 🧐
- Microbiome / Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition 🧐
- Microbiome / 16S Microbial analysis with Nanopore data ✍️ 🧐
- Teaching and Hosting Galaxy training / Motivation and Demotivation 🧐
- Teaching and Hosting Galaxy training / Training techniques to enhance learner participation and engagement 🧐
- Teaching and Hosting Galaxy training / Assessment and feedback in training and teachings 🧐
- Teaching and Hosting Galaxy training / Train-the-Trainer: putting it all together 🧐
- Visualisation / Visualisation with Circos ✍️ 🧐
- Foundations of Data Science / Advanced R in Galaxy 🧐
- Ecology / Metabarcoding/eDNA through Obitools 🧐
- Sequence analysis / NCBI BLAST+ against the MAdLand 🧐
- Sequence analysis / Quality Control 🧐
- Contributing to the Galaxy Training Material / Principles of learning and how they apply to training and teaching ✍️ 🧐
- Contributing to the Galaxy Training Material / Design and plan session, course, materials 🧐
- Contributing to the Galaxy Training Material / Creating content in Markdown 🧐
- Climate / Pangeo ecosystem 101 for everyone - Introduction to Xarray Galaxy Tools 🧐
- Climate / Pangeo Notebook in Galaxy - Introduction to Xarray 🧐
- Epigenetics / Formation of the Super-Structures on the Inactive X 🧐
- Single Cell / Analysis of plant scRNA-Seq Data with Scanpy ✍️
- Single Cell / Pre-processing of 10X Single-Cell ATAC-seq Datasets 🧐
- Single Cell / Pre-processing of 10X Single-Cell RNA Datasets 🧐
- Introduction to Galaxy Analyses / Breve introducción a Galaxy - en español 🧐
Slides
- Development in Galaxy / Galaxy from a developer point of view 🧐
- Variant Analysis / Introduction to Variant analysis 🧐
- Genome Annotation / High Performance Computing for Pairwise Genome Comparison 🧐
- Genome Annotation / Introduction to Genome Annotation 🧐
- Assembly / Unicycler Assembly 🧐
- Assembly / An introduction to get started in genome assembly and annotation 🧐
- Assembly / Genome assembly quality control. 🧐
- Assembly / An Introduction to Genome Assembly 🧐
- Assembly / De Bruijn Graph Assembly 🧐
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads ✍️ 🧐
- Statistics and machine learning / Convolutional neural networks (CNN) Deep Learning - Part 3 🧐
- Statistics and machine learning / Feedforward neural networks (FNN) Deep Learning - Part 1 🧐
- Transcriptomics / Integrate and query local datasets and distant RDF data with AskOmics using Semantic Web technologies 🧐
- Transcriptomics / Introduction to Transcriptomics 🧐
- Transcriptomics / Network Analysis with Heinz 🧐
- Transcriptomics / Identification of non-canonical ORFs and their potential biological function ✍️ 🧐
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana ✍️ 🧐
- Using Galaxy and Managing your Data / Getting data into Galaxy 🧐
- Introduction to Galaxy Analyses / A Short Introduction to Galaxy 🧐
- Introduction to Galaxy Analyses / Introduction to Galaxy 🧐
- Introduction to Galaxy Analyses / Options for using Galaxy 🧐
- Development in Galaxy / Tool Shed: sharing Galaxy tools 🧐
- Development in Galaxy / Generic plugins 🧐
- Development in Galaxy / Tool development and integration into Galaxy 🧐
- Development in Galaxy / Visualizations: JavaScript Plugins 🧐
- Development in Galaxy / Tool Dependencies and Containers 🧐
- Development in Galaxy / Tool Dependencies and Conda 🧐
- Development in Galaxy / Galaxy Interactive Environments 🧐
- Development in Galaxy / Galaxy Interactive Tours 🧐
- Development in Galaxy / Galaxy Webhooks 🧐
-
Imaging
/
Nucleoli Segmentation
&
Feature Extraction
using CellProfiler 🧐 - Metabolomics / Introduction to Metabolomics 🧐
- Metabolomics / Mass spectrometry: LC-MS preprocessing - advanced 🧐
- Proteomics / Introduction to proteomics, protein identification, quantification and statistical modelling 🧐
- Microbiome / Introduction to Microbiome Analysis 🧐
- Microbiome / Introduction to metatranscriptomics 🧐
- Teaching and Hosting Galaxy training / Workshop Kickoff 🧐
- Visualisation / Visualisations in Galaxy 🧐
- Galaxy Server administration / Galaxy from an administrator's point of view 🧐
- Galaxy Server administration / Galaxy on the Cloud 🧐
- Galaxy Server administration / Terraform 🧐
- Galaxy Server administration / Docker and Galaxy 🧐
- Sequence analysis / Quality Control 🧐
- Sequence analysis / Mapping ✍️ 🧐
- Contributing to the Galaxy Training Material / Creating Slides 🧐
- Contributing to the Galaxy Training Material / Contributing with GitHub via command-line 🧐
- Climate / Pangeo ecosystem 101 for everyone 🧐
- Climate / The Pangeo ecosystem 🧐
- Epigenetics / Introduction to DNA Methylation data analysis 🧐
- Epigenetics / Introduction to ATAC-Seq data analysis 🧐
- Epigenetics / Introduction to ChIP-Seq data analysis 🧐
- Epigenetics / ChIP-seq data analysis 🧐
- Epigenetics / EWAS Epigenome-Wide Association Studies Introduction 🧐
- Single Cell / An introduction to scRNA-seq data analysis 🧐
- Introduction to Galaxy Analyses / Una Breve Introducción a Galaxy 🧐
FAQs
- Could I use a different p-adj value for filtering differentially expressed genes?
- Can I use alternative tools for the Quantification step?
Video Recordings
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana 💬
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads 🗣
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana 💬 🗣
- Development in Galaxy / Creating Galaxy tools from Conda Through Deployment 💬 🗣
- Galaxy Server administration / Use Apptainer containers for running Galaxy jobs 💬
GitHub Activity
github Issues Reported
65 Merged Pull Requests
See all of the github Pull Requests and github Commits by Cristóbal Gallardo.
-
 Genome-wide alternative splicing: add slides and fix typos
transcriptomics
Genome-wide alternative splicing: add slides and fix typos
transcriptomics -
 Learning principles training: fix references
contributing
Learning principles training: fix references
contributing -
 Genome-wide alternative splicing training v.2.0
review-neededtranscriptomics
Genome-wide alternative splicing training v.2.0
review-neededtranscriptomics -
 Add as contributor to Learning principles training
contributing
Add as contributor to Learning principles training
contributing -
 Alternative splicing training: improve introduction
transcriptomics
Alternative splicing training: improve introduction
transcriptomics
Reviewed 74 PRs
We love our community reviewing each other's work!
-
 Small updates from feedback from the last workshop
metagenomics
Small updates from feedback from the last workshop
metagenomics -
 Update ChIP-Seq tutorial with latest tool versions and links
epigenetics
Update ChIP-Seq tutorial with latest tool versions and links
epigenetics -
 Genome-wide alternative splicing: add slides and fix typos
transcriptomics
Genome-wide alternative splicing: add slides and fix typos
transcriptomics - github Auto Compress Images
-
 ERGA post-assembly QC training
template-and-toolsassembly
ERGA post-assembly QC training
template-and-toolsassembly
News
New Tutorials: Whole transcriptome analysis of Arabidopsis thaliana
10 April 2021
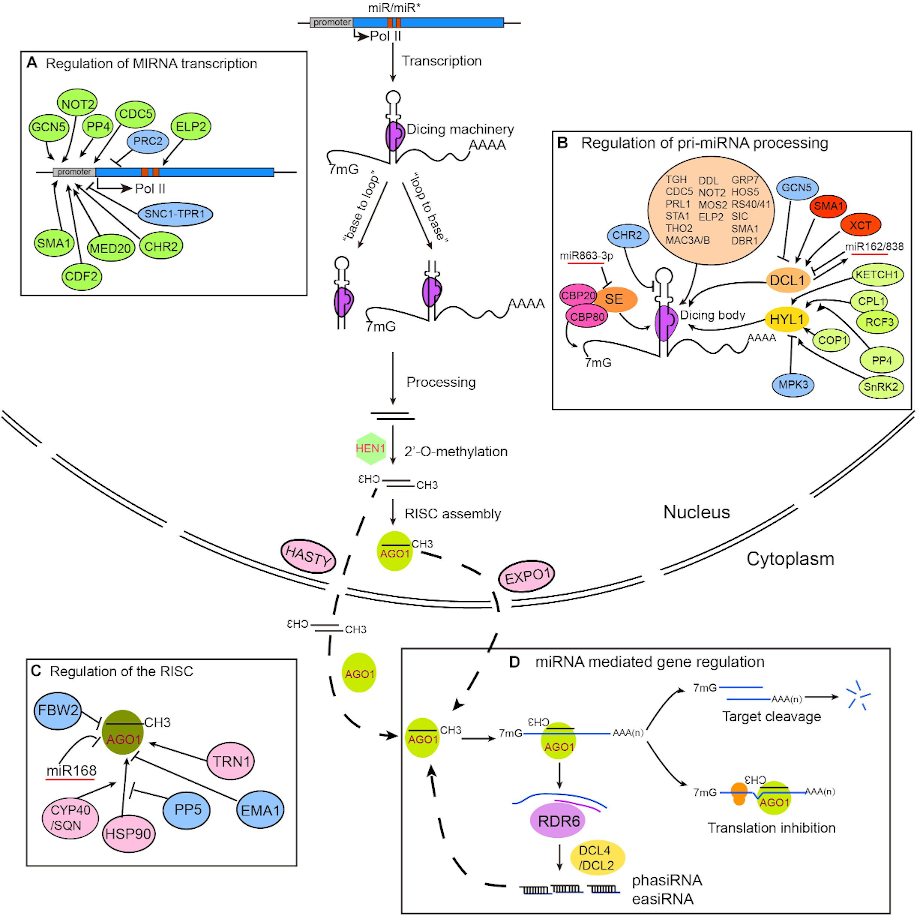
Plant lovers are in luck! A new tutorial on this fascinating kingdom has been added to the GTN. It details the necessary steps to identify potential targets of brassinosteroid-induced miRNAs. Brassinosteroids are phytohormones that have the ability to stimulate plant growth and confer resistance against abiotic and biotic stresses. These characteristics have led to great interest in the field of biotechnology due to its potential for increasing agricultural productivity.
New Tutorial: VGP assembly pipeline
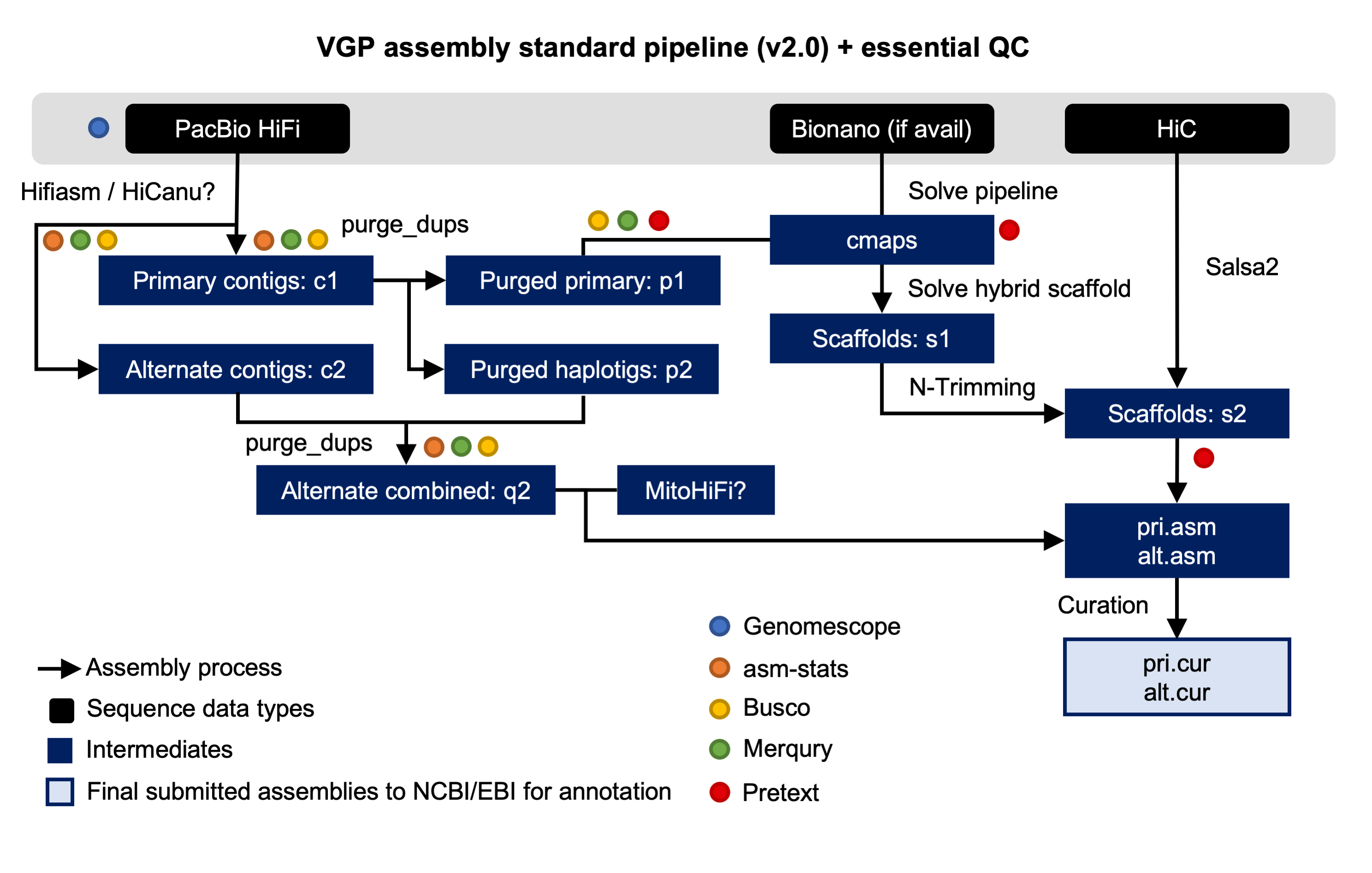
14 March 2022
We are proud to announce that, as result of the collaboration with the Vertebrate Genomes Project (VGP), a new training describing the VGP assembly pipeline is now available in the Galaxy Training Network. The Vertebrate Genomes Project aims to generate high-quality, near-error-free, gap-free, chromosome-level, haplotype-phased, annotated reference genome assemblies for every vertebrate species.