Bérénice Batut
Affiliations
Contributions
The following list includes only slides and tutorials where the individual or organisation has been added to the contributor list. This may not include the sum total of their contributions to the training materials (e.g. GTN css or design, tutorial datasets, workflow development, etc.) unless described by a news post.
Editorial Roles
This contributor has taken on additional responsibilities as an editor for the following topics. They are responsible for ensuring that the content is up to date, accurate, and follows GTN best practices.
- Topic: Contributing to the Galaxy Training Material
- Topic: Foundations of Data Science
- Topic: Development in Galaxy
- Topic: ELIXIR
- Topic: Microbiome
- Topic: Sequence analysis
- Topic: Teaching and Hosting Galaxy training
- Topic: Transcriptomics
- Topic: Variant Analysis
- Learning Pathway: Detection of AMR genes in bacterial genomes
- Learning Pathway: Bacterial comparative genomics
- Learning Pathway: Gallantries Grant - Intellectual Output 2 - Large-scale data analysis, and introduction to visualisation and data modelling
- Learning Pathway: Gallantries Grant - Intellectual Output 4 - Data analysis and modelling for evidence and hypothesis generation and knowledge discovery
- Learning Pathway: Gallantries Grant - Intellectual Output 5 - Train-the-Trainer and mentoring programme
- Learning Pathway: Building and Annotating Metagenome-Assembled Genomes (MAGs) from Metagenomics Reads
- Learning Pathway: Metagenomics data processing and analysis for microbiome
- Learning Pathway: Artificial Intelligence and Machine Learning in Life Sciences using Python
- Learning Pathway: Creation of a Community CoDex and lab (and optionally of a Special Interest Group (SIG))
- Learning Pathway: Train the Trainers
Tutorials
- Computational chemistry / Analysis of molecular dynamics simulations 🧐
- Computational chemistry / Running molecular dynamics simulations using GROMACS 🧐
- Computational chemistry / Running molecular dynamics simulations using NAMD 🧐
- Computational chemistry / Setting up molecular systems 🧐
- Statistics and machine learning / Basics of machine learning 🧐
- Statistics and machine learning / PAPAA PI3K_OG: PanCancer Aberrant Pathway Activity Analysis 🧐
- Statistics and machine learning / Generating Artificial Yeast DNA Sequences using a DNA LLM ✍️ 🧐
- Statistics and machine learning / Age prediction using machine learning 🧐
- Statistics and machine learning / Predicting Mutation Impact with Zero-shot Learning using a pretrained DNA LLM ✍️ 🧐
- Statistics and machine learning / Optimizing DNA Sequences for Biological Functions using a DNA LLM ✍️ 🧐
- Statistics and machine learning / Neural networks using Python 🧐
- Statistics and machine learning / Foundational Aspects of Machine Learning using Python 📝 🧐
- Statistics and machine learning / Machine learning: classification and regression ✍️ 🧐
- Statistics and machine learning / Pretraining a Large Language Model (LLM) from Scratch on DNA Sequences ✍️ 🧐
- Statistics and machine learning / Interval-Wise Testing for omics data 🧐
- Statistics and machine learning / Deep Learning (without Generative Artificial Intelligence) using Python 🧐
- Statistics and machine learning / Fine-tuning a LLM for DNA Sequence Classification ✍️ 🧐
- Statistics and machine learning / Regulations/standards for AI using DOME 📝 🧐
- Galaxy Server administration / Galaxy Tool Management with Ephemeris 🧐
- Galaxy Server administration / Galaxy Installation with Ansible 🧐
- Galaxy Server administration / Connecting Galaxy to a compute cluster 🧐
- Galaxy Server administration / Galaxy Monitoring with gxadmin 🧐
- Galaxy Server administration / Reference Data with CVMFS 🧐
- Galaxy Server administration / Galaxy Monitoring with Telegraf and Grafana 🧐
- Galaxy Server administration / Distributed Object Storage 🧐
- Galaxy Server administration / Data Libraries 🧐
- Galaxy Server administration / Galaxy Monitoring with Reports 🧐
- Galaxy Server administration / Running Jobs on Remote Resources with Pulsar 🧐
- Galaxy Server administration / Ansible 🧐
- Galaxy Server administration / Deploying a compute cluster in OpenStack via Terraform 🧐
- Galaxy Server administration / External Authentication 🧐
- Galaxy Server administration / Galaxy Database schema 🧐
- Galaxy Community Building / Creating a Special Interest Group 🧐
- Galaxy Community Building / Creation of the labs in the different Galaxy instances for your community ✍️ 🧐
- Galaxy Community Building / Creation of resources listing Galaxy workflow for your community ✍️ 🧐
- Galaxy Community Building / Creation of resources listing all the tools and their metadata relevant to your community ✍️ 🧐
- Galaxy Community Building / Creation of a Galaxy tutorial table for your community ✍️ 🧐
- Galaxy Community Building / What's a Special Interest Group? 🧐
- Assembly / Genome Assembly Quality Control 🧐
- Assembly / Genome Assembly of MRSA from Oxford Nanopore MinION data (and optionally Illumina data) ✍️ 🧐
- Assembly / Large genome assembly and polishing 🧐
- Assembly / ERGA post-assembly QC 🧐
- Assembly / An Introduction to Genome Assembly 🧐
- Assembly / Chloroplast genome assembly 🧐
- Assembly / Assembly of the mitochondrial genome from PacBio HiFi reads 🧐
- Assembly / Decontamination of a genome assembly 🧐
- Assembly / Genome Assembly of a bacterial genome (MRSA) sequenced using Illumina MiSeq Data ✍️ 🧐
- Assembly / De Bruijn Graph Assembly 🧐
- Assembly / Unicycler Assembly 🧐
- Assembly / Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads 🧐
- Assembly / Vertebrate genome assembly using HiFi, Bionano and Hi-C data - Step by Step 🧐
- Assembly / Genome assembly using PacBio data 🧐
- Assembly / Using the VGP workflows to assemble a vertebrate genome with HiFi and Hi-C data 🧐
- Assembly / Making sense of a newly assembled genome 🧐
- Genome Annotation / Genome annotation with Maker 🧐
- Genome Annotation / Refining Genome Annotations with Apollo (eukaryotes) 🧐
- Genome Annotation / Genome Annotation 🧐
- Genome Annotation / Identification of AMR genes in an assembled bacterial genome ✍️ 🧐
- Genome Annotation / Creating an Official Gene Set 🧐
- Genome Annotation / Functional annotation of protein sequences 🧐
- Genome Annotation / Essential genes detection with Transposon insertion sequencing ✍️ 🧐
- Genome Annotation / Dataset construction for bacterial comparative genomics ✍️
- Genome Annotation / Genome annotation with Funannotate 🧐
- Genome Annotation / Masking repeats with RepeatMasker 🧐
- Genome Annotation / Bacterial genome quality control ✍️
- Genome Annotation / Genome annotation with Maker (short) 🧐
- Genome Annotation / Refining Genome Annotations with Apollo (prokaryotes) 🧐
- Genome Annotation / Genome annotation with Braker3 🧐
- Genome Annotation / Bacterial pangenomics ✍️
- Genome Annotation / Genome annotation with Helixer 🧐
- Genome Annotation / Bacterial Genome Annotation ✍️ 🧐
- Genome Annotation / Genome annotation with Prokka 🧐
- Genome Annotation / Comparative gene analysis in unannotated genomes 🧐
- Using Galaxy and Managing your Data / Group tags for complex experimental designs 🧐
- Using Galaxy and Managing your Data / Automating Galaxy workflows using the command line 🧐
- Using Galaxy and Managing your Data / RStudio in Galaxy ✍️ 🧐
- Using Galaxy and Managing your Data / Extracting Workflows from Histories 🧐
- Using Galaxy and Managing your Data / Use Jupyter notebooks in Galaxy 🧐
- Using Galaxy and Managing your Data / Rule Based Uploader: Advanced 🧐
- Using Galaxy and Managing your Data / InterMine integration with Galaxy 🧐
- Using Galaxy and Managing your Data / Using dataset collections 🧐
- Using Galaxy and Managing your Data / Annotate, prepare tests and publish Galaxy workflows in workflow registries ✍️ 🧐
- Using Galaxy and Managing your Data / Name tags for following complex histories 🧐
- Using Galaxy and Managing your Data / Understanding Galaxy history system 📝 🧐
- Ecology / RAD-Seq to construct genetic maps 🧐
- Ecology / Species distribution modeling ✍️
- Ecology / RAD-Seq de-novo data analysis 🧐
- Ecology / Regional GAM ✍️
- Ecology / Preparing genomic data for phylogeny reconstruction 🧐
- Ecology / Checking expected species and contamination in bacterial isolate ✍️
- Ecology / RAD-Seq Reference-based data analysis 🧐
- Evolution / Phylogenetic analysis for bacterial comparative genomics ✍️
- Evolution / Tree thinking for tuberculosis evolution and epidemiology 🧐
- Metabolomics / Mass spectrometry imaging: Examining the spatial distribution of analytes 🧐
- Metabolomics / Mass spectrometry imaging: Finding differential analytes 🧐
- Microbiome / Antibiotic resistance detection 🧐
- Microbiome / Remove contamination and host reads ✍️
- Microbiome / QIIME 2 Cancer Microbiome Intervention 🧐
- Microbiome / Building and Annotating Metagenome-Assembled Genomes (MAGs) from Short Metagenomics Paired Reads ✍️ 🧐
- Microbiome / Analyses of metagenomics data - The global picture ✍️ 🧐
- Microbiome / Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition ✍️ 🧐
- Microbiome / Calculating α and β diversity from microbiome taxonomic data ✍️ 🧐
- Microbiome / Taxonomic Profiling and Visualization of Metagenomic Data ✍️ 🧐
- Microbiome / Assembly of metagenomic sequencing data ✍️ 🧐
- Microbiome / Building an amplicon sequence variant (ASV) table from 16S data using DADA2 ✍️ 🧐
- Microbiome / Binning of metagenomic sequencing data 📝 🧐
- Microbiome / 16S Microbial analysis with Nanopore data 🧐
- Microbiome / Identification of the micro-organisms in a beer using Nanopore sequencing ✍️ 🧐
- Microbiome / Query an annotated mobile genetic element database to identify and annotate genetic elements (e.g. plasmids) in metagenomics data ✍️ 🧐
- Microbiome / 16S Microbial Analysis with mothur (short) ✍️ 🧐
- Microbiome / QIIME 2 Moving Pictures 🧐
- Microbiome / 16S Microbial Analysis with mothur (extended) ✍️ 🧐
- Microbiome / Metatranscriptomics analysis using microbiome RNA-seq data (short) ✍️ 🧐
- Microbiome / Metatranscriptomics analysis using microbiome RNA-seq data ✍️ 🧐
- Variant Analysis / Mapping and molecular identification of phenotype-causing mutations 🧐
- Variant Analysis / Identification of somatic and germline variants from tumor and normal sample pairs 🧐
- Variant Analysis / M. tuberculosis Variant Analysis 🧐
- Variant Analysis / Calling variants in diploid systems 🧐
- Variant Analysis / Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data ✍️ 🧐
- Variant Analysis / Microbial Variant Calling 🧐
- Variant Analysis / Calling variants in non-diploid systems 🧐
- Variant Analysis / Calling very rare variants 🧐
- Variant Analysis / Exome sequencing data analysis for diagnosing a genetic disease ✍️ 🧐
- Transcriptomics / RNA Seq Counts to Viz in R ✍️ 🧐
- Transcriptomics / CLIP-Seq data analysis from pre-processing to motif detection ✍️ 🧐
- Transcriptomics / Reference-based RNAseq data analysis (long) 🧐
- Transcriptomics / Reference-based RNA-Seq data analysis ✍️ 🧐
- Transcriptomics / 1: RNA-Seq reads to counts 🧐
- Transcriptomics / Whole transcriptome analysis of Arabidopsis thaliana 🧐
- Transcriptomics / Differential abundance testing of small RNAs 🧐
- Transcriptomics / Visualization of RNA-Seq results with CummeRbund 🧐
- Transcriptomics / Network analysis with Heinz 🧐
- Transcriptomics / De novo transcriptome reconstruction with RNA-Seq 🧐
- Foundations of Data Science / Python - Warm-up for statistics and machine learning 📝 🧐
- Foundations of Data Science / Advanced R in Galaxy ✍️
- Foundations of Data Science / One protein along the UniProt page ✍️ 🧐
- Foundations of Data Science / Make & Snakemake 🧐
- Foundations of Data Science / Learning about one gene across biological resources and formats ✍️ 🧐
- Foundations of Data Science / R basics in Galaxy 📝 🧐
- Foundations of Data Science / Advanced SQL 🧐
- Contributing to the Galaxy Training Material / Principles of learning and how they apply to training and teaching ✍️ 🧐
- Contributing to the Galaxy Training Material / Contributing to the Galaxy Training Network with GitHub ✍️ 🧐
- Contributing to the Galaxy Training Material / Creating a new tutorial ✍️ 🧐
- Contributing to the Galaxy Training Material / Adding auto-generated video to your slides 🧐
- Contributing to the Galaxy Training Material / Contributing with GitHub via its interface ✍️ 🧐
- Contributing to the Galaxy Training Material / Generating PDF artefacts of the website ✍️ 🧐
- Contributing to the Galaxy Training Material / Preview the GTN website as you edit your training material ✍️
- Contributing to the Galaxy Training Material / Creating content in Markdown ✍️ 🧐
- Contributing to the Galaxy Training Material / Creating Interactive Galaxy Tours ✍️ 🧐
- Contributing to the Galaxy Training Material / Tools, Data, and Workflows for tutorials ✍️ 🧐
- Contributing to the Galaxy Training Material / Including a new topic ✍️ 🧐
- Contributing to the Galaxy Training Material / Updating tool versions in a tutorial 📝 🧐
- Contributing to the Galaxy Training Material / Design and plan session, course, materials ✍️ 🧐
- Contributing to the Galaxy Training Material / FAIR-by-Design methodology 🧐
- Epigenetics / Hi-C analysis of Drosophila melanogaster cells using HiCExplorer 🧐
- Epigenetics / Identification of the binding sites of the Estrogen receptor ✍️ 🧐
- Epigenetics / Identification of the binding sites of the T-cell acute lymphocytic leukemia protein 1 (TAL1) 🧐
- Epigenetics / Infinium Human Methylation BeadChip 🧐
- Epigenetics / Formation of the Super-Structures on the Inactive X ✍️ 🧐
- Epigenetics / DNA Methylation data analysis 🧐
- Climate / Functionally Assembled Terrestrial Ecosystem Simulator (FATES) 🧐
- Single Cell / Clustering 3K PBMCs with Scanpy ✍️
- Single Cell / Combining single cell datasets after pre-processing 🧐
- Single Cell / Pre-processing of 10X Single-Cell RNA Datasets 🧐
- Single Cell / Pre-processing of Single-Cell RNA Data ✍️ 🧐
- Single Cell / Understanding Barcodes 🧐
- Single Cell / Downstream Single-cell RNA analysis with RaceID 🧐
- Introduction to Galaxy Analyses / IGV Introduction 🧐
- Introduction to Galaxy Analyses / NGS data logistics 🧐
- Introduction to Galaxy Analyses / A short introduction to Galaxy 📝 🧐
- Introduction to Galaxy Analyses / Galaxy Basics for everyone 🧐
- Introduction to Galaxy Analyses / Upload data to Galaxy ✍️ 🧐
- Introduction to Galaxy Analyses / From peaks to genes ✍️ 🧐
- Introduction to Galaxy Analyses / Introduction to Genomics and Galaxy 🧐
- Introduction to Galaxy Analyses / Very Short Introductions: QC 🧐
- Introduction to Galaxy Analyses / Galaxy Basics for genomics 🧐
- Development in Galaxy / Data source integration ✍️ 🧐
- Development in Galaxy / JavaScript plugins 🧐
- Development in Galaxy / Adding and updating best practice metadata for Galaxy tools using the bio.tools registry ✍️ 🧐
- Development in Galaxy / Generic plugins 🧐
- Development in Galaxy / Galaxy Webhooks 🧐
- Proteomics / metaQuantome 3: Taxonomy 🧐
- Proteomics / Clinical Metaproteomics 2: Discovery 🧐
- Proteomics / Detection and quantitation of N-termini (degradomics) via N-TAILS 🧐
- Proteomics / Clinical Metaproteomics 4: Quantitation 🧐
- Proteomics / Mass spectrometry imaging: Loading and exploring MSI data 🧐
- Proteomics / Protein FASTA Database Handling 🧐
- Proteomics / Annotating a protein list identified by LC-MS/MS experiments 🧐
- Proteomics / Peptide and Protein ID using SearchGUI and PeptideShaker 🧐
- Proteomics / Peptide and Protein ID using OpenMS tools 🧐
- Proteomics / Clinical Metaproteomics 5: Data Interpretation 🧐
- Proteomics / Metaproteomics tutorial 🧐
- Proteomics / Clinical Metaproteomics 3: Verification 🧐
- Proteomics / Peptide and Protein Quantification via Stable Isotope Labelling (SIL) 🧐
- Proteomics / Label-free versus Labelled - How to Choose Your Quantitation Method 🧐
- Proteomics / Secretome Prediction 🧐
- Proteomics / metaQuantome 2: Function 🧐
- Proteomics / metaQuantome 1: Data creation 🧐
- Proteomics / Clinical Metaproteomics 1: Database-Generation 🧐
- Visualisation / Genomic Data Visualisation with JBrowse 🧐
- Teaching and Hosting Galaxy training / Teaching experiences ✍️ 🧐
- Teaching and Hosting Galaxy training / Teaching online ✍️
- Teaching and Hosting Galaxy training / Galaxy Admin Training 🧐
- Teaching and Hosting Galaxy training / Motivation and Demotivation ✍️ 🧐
- Teaching and Hosting Galaxy training / Hybrid training ✍️
- Teaching and Hosting Galaxy training / Organizing a workshop ✍️
- Teaching and Hosting Galaxy training / Asynchronous training ✍️
- Teaching and Hosting Galaxy training / Running a workshop as an instructor ✍️
- Teaching and Hosting Galaxy training / Set up a Galaxy for Training ✍️ 🧐
- Teaching and Hosting Galaxy training / Live Coding is a Skill ✍️
- Teaching and Hosting Galaxy training / Train-the-Trainer: putting it all together ✍️ 🧐
- Teaching and Hosting Galaxy training / Training techniques to enhance learner participation and engagement ✍️ 🧐
- Teaching and Hosting Galaxy training / Assessment and feedback in training and teachings ✍️ 🧐
- Sequence analysis / Mapping ✍️ 🧐
- Sequence analysis / SARS-CoV-2 Viral Sample Alignment and Variant Visualization 🧐
- Sequence analysis / Removal of human reads from SARS-CoV-2 sequencing data 🧐
- Sequence analysis / Quality Control ✍️ 🧐
- Sequence analysis / Quality and contamination control in bacterial isolate using Illumina MiSeq Data ✍️ 🧐
Slides
- Development in Galaxy / Galaxy from a developer point of view ✍️ 🧐
- Statistics and machine learning / Neural networks using Python 🧐
- Statistics and machine learning / Deep Learning (without Generative Artificial Intelligence) using Python 🧐
- Statistics and machine learning / Regulations/standards for AI using DOME 🧐
- Galaxy Server administration / Connecting Galaxy to a compute cluster 🧐
- Galaxy Server administration / User, Role, Group, Quota, and Authentication managment 🧐
- Galaxy Server administration / Docker and Galaxy ✍️ 🧐
- Galaxy Server administration / Galaxy from an administrator's point of view 🧐
- Galaxy Server administration / Server: Other 🧐
- Galaxy Server administration / Advanced customisation of a Galaxy instance 🧐
- Assembly / An Introduction to Genome Assembly 🧐
- Assembly / De Bruijn Graph Assembly 🧐
- Assembly / Unicycler Assembly 🧐
- Genome Annotation / Essential genes detection with Transposon insertion sequencing 🧐
- Genome Annotation / Introduction to Genome Annotation 🧐
- Genome Annotation / Genome annotation with Prokka 🧐
- Using Galaxy and Managing your Data / Getting data into Galaxy 🧐
- Metabolomics / Introduction to Metabolomics 🧐
- Microbiome / Introduction to Microbiome Analysis ✍️ 🧐
- Variant Analysis / Introduction to Variant analysis ✍️ 🧐
- Transcriptomics / Introduction to Transcriptomics ✍️ 🧐
- Transcriptomics / Visualization of RNA-Seq results with CummeRbund 🧐
- Foundations of Data Science / Bioinformatics Data Types and Databases 🧐
- Contributing to the Galaxy Training Material / Contributing with GitHub via command-line ✍️ 🧐
- Contributing to the Galaxy Training Material / Creating Slides ✍️ 🧐
- Contributing to the Galaxy Training Material / Overview of the Galaxy Training Material ✍️ 🧐
- Epigenetics / Introduction to ChIP-Seq data analysis ✍️ 🧐
- Epigenetics / EWAS Epigenome-Wide Association Studies Introduction 🧐
- Epigenetics / ChIP-seq data analysis ✍️ 🧐
- Epigenetics / Introduction to DNA Methylation data analysis 🧐
- Climate / Functionally Assembled Terrestrial Ecosystem Simulator (FATES) 🧐
- Climate / Introduction to climate data 🧐
- Introduction to Galaxy Analyses / A Short Introduction to Galaxy 🧐
- Introduction to Galaxy Analyses / Introduction to Galaxy ✍️ 🧐
- Introduction to Galaxy Analyses / Options for using Galaxy 🧐
- Development in Galaxy / Galaxy Interactive Environments 🧐
- Development in Galaxy / Scripting Galaxy using the API and BioBlend 🧐
- Development in Galaxy / Visualizations: JavaScript Plugins 🧐
- Development in Galaxy / Tool Shed: sharing Galaxy tools ✍️ 🧐
- Development in Galaxy / Tool Dependencies and Containers 🧐
- Development in Galaxy / Prerequisites for building software/conda packages 🧐
- Development in Galaxy / Generic plugins 🧐
- Development in Galaxy / Galaxy Webhooks 🧐
- Development in Galaxy / Tool Dependencies and Conda 🧐
- Development in Galaxy / Tool development and integration into Galaxy ✍️ 🧐
- Development in Galaxy / Galaxy Interactive Tours ✍️ 🧐
- Proteomics / Introduction to proteomics, protein identification, quantification and statistical modelling 🧐
- Visualisation / Visualisations in Galaxy 🧐
- Sequence analysis / Mapping 🧐
- Sequence analysis / Quality Control ✍️ 🧐
FAQs
- Average Nucleotide Identity (ANI): A Measure of Genomic Similarity
- ANI threshold for dereplication
- CheckM2 vs CheckM
- How do I add my community to the Galaxy CoDex?
- Thanks!
- How can I get started with contributing?
- What is a Codex?
- Adding a tag
- Changing the datatype
- Creating a new file
- Detecting the datatype (file format)
- Importing data from a data library
- Importing data from remote files
- Importing via links
- Renaming a dataset
- Upload few files (1-10)
- Upload many files (>10) via FTP
- The tutorial uses the normalised count table for visualisation. What about using VST normalised counts or rlog normalised counts?
- FASTQ format
- What is Galaxy?
- GTN ADR: Image Storage
- GTN ADR: Why Jekyll and not another Static Site Generator (SSG)
- GTN Architectural Decision Record Template
- What is this website?
- How can I advertise the training materials on my posters?
- What audiences are the tutorials for?
- How can I cite the GTN?
- How is the content licensed?
- Sustainability of the training-material and metadata
- What are the tutorials for?
- Copy a dataset between histories
- Créer un nouvel history
- Creating a new history
- Importing a history
- Renaming a history
- When is the "infer experiment" tool used in practice?
- What are the best practices for teaching with Galaxy?
- What Galaxy instance should I use for my training?
- How do I get help?
- Where do I start?
- Launch RStudio
- Kraken2 and the k-mer approach for taxonomy classification
- How can I get help?
- Where do I start?
- Where can I run the hands-on tutorials?
- How do I use this material?
- Maximum MAG contamination percentage
- Minimum MAG completeness percentage
- Minimum MAG length
- Getting your API key
- Quality Scores
- Is it possible to visualize the RNA STAR bam file using the JBrowse tool?
- In 'infer experiments' I get unequal numbers, but in the IGV it looks like it is unstranded. What does this mean?
- What is Taxonomy?
- Selecting a dataset collection as input
- Select multiple datasets
- Add genome and annotations to IGV from Galaxy
- Add Mapped reads track to IGV from Galaxy
- Annotate a workflow
- Get the workflow invocation
- Importing a workflow using the Tool Registry Server (TRS) search
- Make a workflow public
- Running a workflow
Video Recordings
- Genome Annotation / Bacterial Genome Annotation 💬 🗣
- Microbiome / Antibiotic resistance detection 💬
- Microbiome / Taxonomic Profiling and Visualization of Metagenomic Data 🗣
- Microbiome / Assembly of metagenomic sequencing data 💬 🗣
- Microbiome / Identification of the micro-organisms in a beer using Nanopore sequencing 🗣
- Transcriptomics / Reference-based RNA-Seq data analysis 🗣
- Contributing to the Galaxy Training Material / Creating a new tutorial 🗣
- Sequence analysis / Quality Control 💬
Events
- My Training Event Title 🧑🏫
- Galaxy Training Academy 2025 🧑🏫
- Galaxy Beyond Basics: Mastering Workflows, Automation, and Scalability 🎪 🧑🏫
- Galaxy Training Academy 2024 🧑🏫
GitHub Activity
github Issues Reported
314 Merged Pull Requests
See all of the github Pull Requests and github Commits by Bérénice Batut.
-
 [Microbiome] Add 2 modules to MAGs learning pathway
template-and-tools
[Microbiome] Add 2 modules to MAGs learning pathway
template-and-tools -
 [Microbiome] Add tutorial for MAGs building and annotation from raw reads
faqsmicrobiome
[Microbiome] Add tutorial for MAGs building and annotation from raw reads
faqsmicrobiome -
 [Microbiome] Add learning pathway for MAGs building and annotation
template-and-tools
[Microbiome] Add learning pathway for MAGs building and annotation
template-and-tools -
 Add Lisanna as contributor to 2025-07-01-gtn-elixir-life-cycle.md
news
Add Lisanna as contributor to 2025-07-01-gtn-elixir-life-cycle.md
news -
 Add news about GTN and ELIXIR Training Life-Cycle
news
Add news about GTN and ELIXIR Training Life-Cycle
news
Reviewed 432 PRs
We love our community reviewing each other's work!
-
 [Microbiome] Add tutorial for MAGs building and annotation from raw reads
faqsmicrobiome
[Microbiome] Add tutorial for MAGs building and annotation from raw reads
faqsmicrobiome -
 Galaxy Formation Avancée 2026
template-and-tools
Galaxy Formation Avancée 2026
template-and-tools -
 Minor fix for metagenomic assembly tutorial
microbiome
Minor fix for metagenomic assembly tutorial
microbiome -
 Update metagenomics assembly tutorial
microbiome
Update metagenomics assembly tutorial
microbiome -
 [Microbiome] Add learning pathway for MAGs building and annotation
template-and-tools
[Microbiome] Add learning pathway for MAGs building and annotation
template-and-tools
News
Next GTN CoFest May 20, 2021
18 March 2021
Every 3 months we organise one day dedicated to the GTN community! Thursday, May 20th is the next GTN CoFest. We will be working on the training materials, have discussions with the global Galaxy traning community. Do you teach with Galaxy? Want to learn how to add your tutorial to the GTN? New to the community and just want to learn more? Everybody is welcome!
New Feature: FAQs
24 March 2021
Snippets have received an major overhaul! Snippets/FAQs are small reusable bits of training, often answering a single question. In addition to the general Galaxy snippets, we now also support topic-level and tutorial-level FAQs. This will allow contributors to add common questions and answers to their tutorials, which not only can be included in the tutorial text itself, but are also used to autogenerate an FAQ page for the tutorial or topic listing all common questions (and answers).This will be useful both to participants following tutorials via self-study, and also to instructors preparing for a training event.
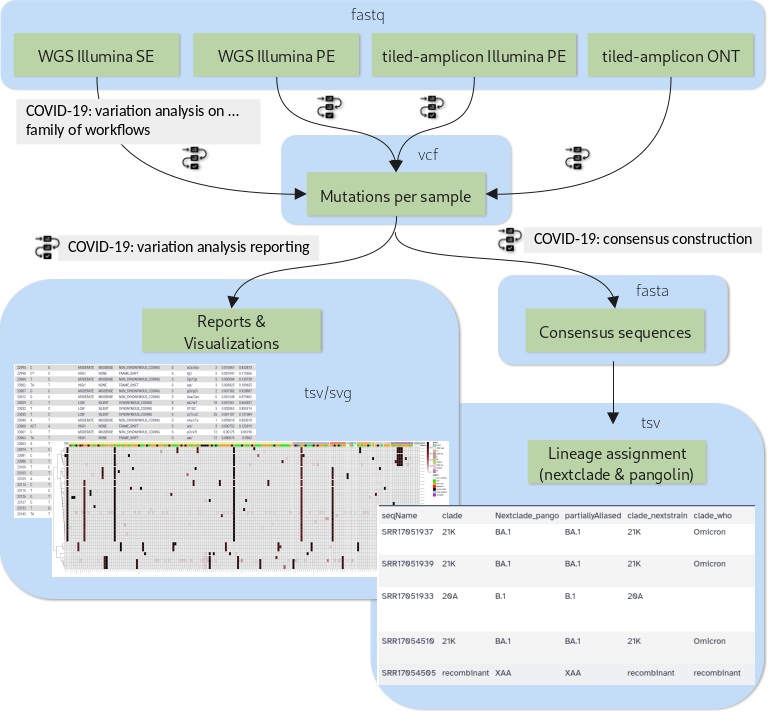
New Tutorial: Mutation calling, viral genome reconstruction and lineage/clade assignment from SARS-CoV-2 sequencing data
30 June 2021
Effectively monitoring global infectious disease crises, such as the COVID-19 pandemic, requires capacity to generate and analyze large volumes of sequencing data in near real time. These data have proven essential for monitoring the emergence and spread of new variants, and for understanding the evolutionary dynamics of the virus.
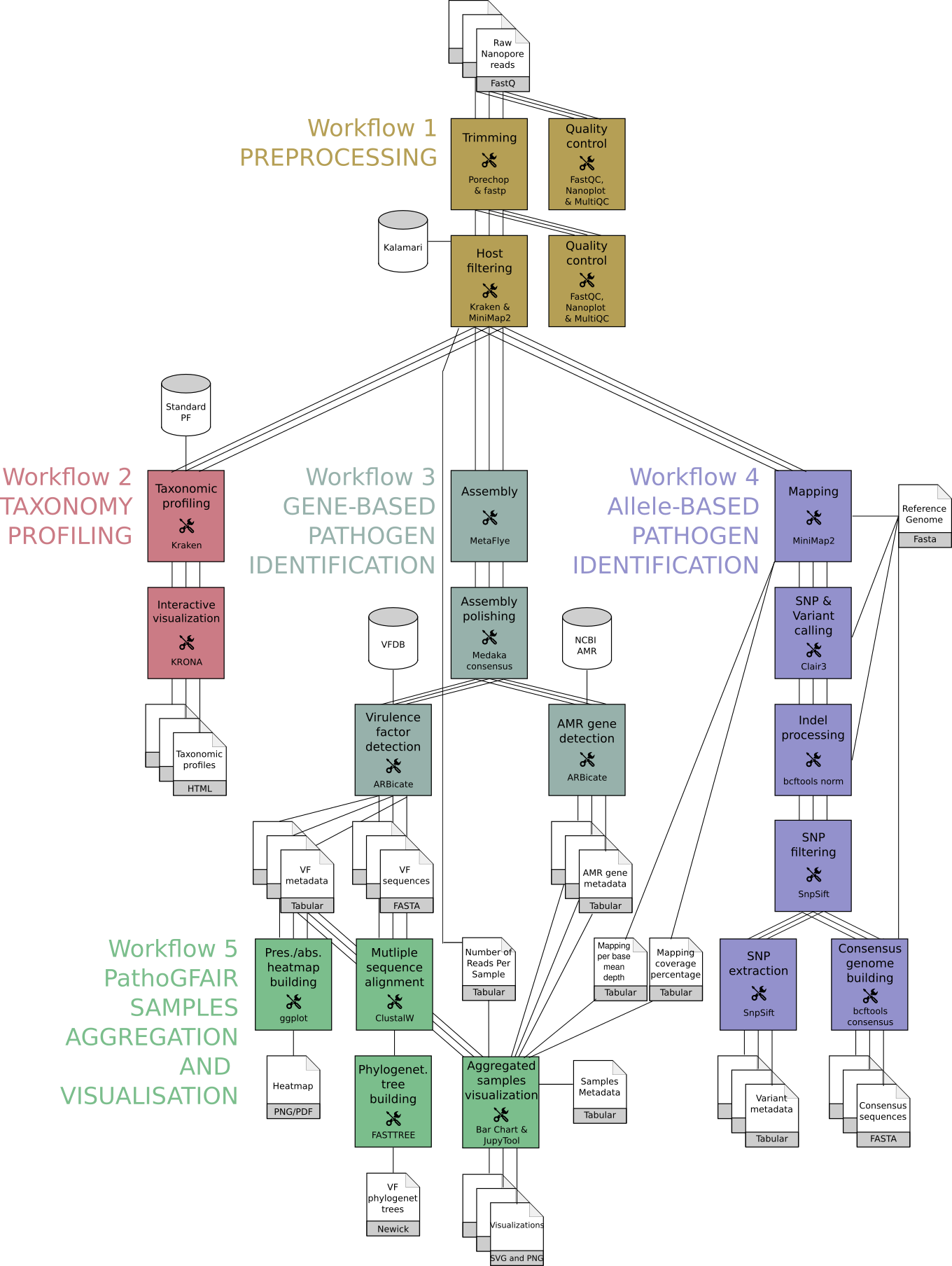
New Tutorial: Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition
26 January 2023
Food contamination with pathogens are a major burden on our society. Globally, they affect an estimated 600 million people a year and impact socioeconomic development at different levels. These outbreaks are mainly due to Salmonella spp. followed by Campylobacter spp. and Noroviruses.
Enhancing Scientific Training: The Galaxy Training Network's Role in the ELIXIR Training Life-Cycle
1 July 2025
In the rapidly evolving landscape of data science, continuous learning and skill development are crucial. The Galaxy Training Network (GTN) plays a pivotal role in this educational ecosystem, particularly within the ELIXIR Training Life-Cycle. This blog post explores how the GTN contributes to each phase of the life-cycle and aligns with the SPLASH recommendations, ensuring high-quality training for researchers worldwide.
External Links

Favourite Topics
Favourite Formats